Abstract
The MazEF systems are thought to contribute to the capacity for long-term dormancy observed in the human pathogen, Mycobacterium tuberculosis. However, except for their functions as mRNA interferases, little is known regarding any additional cellular functions of these systems in the pathogen. In the present study, we observed a negative interplay between MazF protein Rv1495 and the sole M. tuberculosis DNA topoisomerase I (MtbTopA) with respect to protein functions. Through its C-terminal domain, MtbTopA physically interacted with and inhibited the mRNA cleavage activity of Rv1495. Rv1495, in turn, inhibited the DNA cleavage activity of MtbTopA as well as its function of relaxation of supercoiled DNA. An N-terminus fragment of Rv1495, designated Rv1495-N(29-56), lost mRNA cleavage activity, but retained a significant physical interaction and inhibitory effect on TopA proteins from both M. tuberculosis and M. smegmatis. This fragment, although less effective than the full-length protein, was able to inhibit mycobacterial growth when expressed through a recombinant plasmid in M. smegmatis. The Rv1495 physically interacted with the M. smegmatis TopA both in vitro and in vivo. Our findings imply that MazEF systems can affect bacterial survival by a novel mechanism that allows direct modulation of M. tuberculosis topoisomerase I.
INTRODUCTION
Toxin–antitoxin (TA) gene cassettes are widely found in many free-living prokaryotes including a number of pathogenic bacteria such as Mycobacterium tuberculosis (1,2). These gene cassettes appear to be involved in bacterial survival as well as in the formation of persistent pathogenic infections in hostile host-cell environments (3). In a typical TA system, the toxin gene is usually co-expressed with its cognate antitoxin gene present in the same operon (4,5). The Escherichia coli chromosome, for example, contains six such operons, two of which [the relBE (6) and the mazEF (1) systems] have been most extensively studied (5). The mazEF system consists of two adjacent genes, mazE and mazF, and has the properties required for an addiction module. MazF is stable and toxic, whereas MazE is a labile and antitoxic protein (1). MazE and MazF interact with each other to form a complex, MazE–MazF, which negatively regulates the expression of mazEF (7–9). Several recent studies have revealed MazF to be a sequence-specific endoribonuclease that recognizes specific cleavage sites within single-strand RNA. For this reason, it has been termed an mRNA interferase (8,10,11).
Mycobacterium tuberculosis is one of the most devastating human pathogens, causing the deaths of about two million people each year around the globe. As it infects, this pathogen confronts a particularly hostile host cell environment, including a restricted access to nutrients and a reduced oxygen tension within the host macrophage cells (12–14). The presence of TA systems in M. tuberculosis may contribute to its ability to undergo long term dormancy within these inhospitable human tissues, and consequently might prove to be essential for the persistence of the pathogen (3). This potential importance of TA systems in the life cycle of M. tuberculosis is suggested by the presence of a remarkably large number of these TA genes in its genome (15). Of the nine MazF homologs discovered thus far in M. tuberculosis (2,3,10,16), four have been shown to cause cell growth arrest when induced in E. coli. The bacterial MazF also shows an mRNA interferase activity both in vivo and in vitro (3,10,16). However, except for this function as an interferase, little is known about cellular effects of the other numerous toxins in this pathogen.
In contrast to its large number of TA genes, the genome of M. tuberculosis encodes only one single topoisomerase I (MtbTopA) (15). A related but nonpathogenic organism, M. smegmatis, also possesses a single TopA-homolog (MsmTopA) (17) that features a conserved N-terminal domain (NTD) and a highly variable C-terminal domain (CTD) (18). The MtbTopA topoisomerase might be essential for preventing hypernegative supercoiling of DNA during transcription (19), and may also be involved in pathogen survival as attempts to isolate transposon insertion mutants in the topA gene have thus far been unsuccessful (20). Although both the MazEF system and DNA topoisomerase I appear to function in bacterial stress responses (3,20), interaction between these two proteins remains to be characterized.
Recently, the M. tuberculosis MazF homolog, Rv1495, was characterized as a toxin with ribonuclease activity (3,10). In the current study, we report on an interplay between Rv1495 proteins and DNA topoisomerase I that results in mutual inhibition of their activities. An N-terminus fragment of Rv1495, Rv1495-N(29-56), loses its mRNA cleavage activity but retains its ability to physically interact with and inhibit TopA from both M. tuberculosis and M. smegmatis. The Rv1495-N(29-56) fragment is further shown to inhibit mycobacterial growth. These findings suggest a new mechanism that allows the pathogenic MazEF systems of M. tuberculosis to participate in modulating the activities of DNA topoisomerase I.
MATERIALS AND METHODS
DNA primers and peptides
All DNA primers for polymerase chain reactions (PCR) were synthesized by Invitrogen and are listed in Supplementary Table S1. Two short peptides Rv1495-N28: MNAPLRGQ VYRCDLGYGAKPWL IVSNNA, and Rv1495-N(29-56): RNRHTADVVAVRLTTTRRTIPTWVAMGP were synthesized by GL Biochem (Shanghai) Ltd.
Bacterial strains, plasmids, enzymes and chemicals
The host strain E. coli BL21 and pET28a vector (Novagen) were used to express M. tuberculosis proteins. The plasmids pBT, pTRG and E. coli reporter strains for the bacterial two-hybrid assays were purchased from Stratagene. pBluescript and pGEX-4T-1 were purchased from Pharmacia. Restriction enzymes, T4 DNA ligase, DNA polymerase, modification enzymes, deoxynucleoside triphosphates (dNTPs) and all antibiotics were purchased from TaKaRa Biotech. DNA purification kits were purchased from Watson Biotechnologies. All plasmids constructed in this study are listed in Supplementary Table S2.
Cloning, expression and purification of M. tuberculosis proteins
Rv1495 and the topoisomerase I gene from M. tuberculosis or M. smegmatis genome were amplified using their PCR primers (Supplementary Table S1) and cloned into the prokaryotic expression vector pET28a or pGEX-4T-1. Escherichia coli BL21 (Novagen) was used as the host strain to express and purified the proteins according to our previously published procedures (21–23). Both 6× his tagged and GST-fused recombinant proteins were prepared for activity and protein–protein interaction assays. Purified proteins were >95% pure as determined by SDS–PAGE and subsequent staining by Coomassie blue. Protein concentrations were determined by spectrophotometric absorbance at 260 nm according to Gill and von Hippel (24).
Bacterial two-hybrid assay
The BacterioMatch II Two-Hybrid System Library Construction Kit (Stratagene) was used to detect protein–protein interactions between Rv1495 and topoisomerase I proteins. Bacterial two-hybrid system detects protein–protein interactions based on transcriptional activation and analysis was carried out according to previously published procedures (21–23). Positive growth cotransformants were selected on the Selective Screening Medium plate containing 5 mM 3-amino-1,2,4-triazole (3-AT) (Stratagene), 8 μg/ml streptomycin, 15 μg/ml tetracycline, 34 μg/ml chloramphenicol and 50 μg/ml kanamycin. Cotransformants containing pBT-LGF2 and pTRG-Gal11P (Stratagene) were used as positive controls for an expected growth on the Screening Medium. Cotransformants containing empty vector pBT and pTRG were used as negative controls.
DNA relaxation assay
The assay was carried out according to the previously published procedure (25,26). The supercoiled pBluescript was incubated in the presence of mycobacterial topoisomerase I (0–0.15 μM) with or without various amounts of Rv1495 or its mutant proteins, and incubated at 37°C for 40 min. After incubation, proteinase K (1 mg/ml) and its buffer (100 mM Tris–HCl, pH 7.8, 5 mM EDTA, 0.5% SDS) were added to the mixture and were further co-incubated for 15 min at 37°C. Samples were resolved by 0.9% agarose gel electrophoresis and then stained with ethidium bromide for analysis. Each analysis was performed at least in triplicate and a representative set of data was presented.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was carried out to analyze single-stranded DNA-binding activity of MtbTopA, as described earlier (27). 5′-end-labeled 32-mer oligonucleotide (5′-CAGTGAGCGAGCTTCCGCTTGACATCCC AATA-3′) (27) was used as a specific single-stranded DNA substrate. The reaction mixtures containing 40 mM Tris–HCl (pH 8.0), 20 mM NaCl, 1 mM EDTA, 150 nM oligonucleotide were incubated with varying concentrations of MtbtopA (60–240 nM) or Rv1495(240–960 nM) for 40 min on ice. Rv0054 (single-stranded DNA-binding protein) was used as a positive control. The protein–DNA complexes were resolved on 6% non-denaturing PAGE (39.5:0.5) using 0.5× TBE (Tris–borate–EDTA) as the running buffer. Gels were exposed to a storage-phosphor screen overnight at room temperature. The images were acquired by a Typhoon Scanner (GE healthcare). EMSA with plasmid pBluescript DNA was carried out to analyze the DNA-binding activity of Rv1495. The reactions contained 40 mM Tris–HCl (pH 8.0), 20 mM NaCl, 1 mM EDTA and 1.5–6 μM protein. Reactions were performed on ice for 30 min. Samples were resolved by 0.8% TBE agarose gel electrophoresis and then stained with ethidium bromide for analysis.
Oligonucleotide cleavage assay
The assays for the single-stranded DNA-binding cleavage activity of MtbTopA were performed as described earlier (27). Varying concentrations of MtbTopA (100–400 nM) were co-incubated at 37°C for 30 min with 200 nM 32-mer oligonucleotide, in the same buffer used for EMSA to yield a 5′-end-labeled 19-mer cleaved product. The effects of Rv1495 on the cleavage activity of MtbTopA (0.2 μM) were analyzed using varying concentrations of Rv1495 (3–9 μM). Heat-denatured Rv1495 and GST proteins were used as negative controls. The reactions were stopped by adding 10 μl loading buffer (95% formamide, 20 mM EDTA, 0.05% bromphenol blue and 0.05% xylene cyanol FF). The samples were incubated at 90°C for 5 min prior to electrophoresis on a 20% polyacrylamide and 7 M urea gel. The images were acquired by a Typhoon Scanner (GE Healthcare).
Assay for toxin growth inhibition
A TetR-controlled expression system was used to analyze the effects of Rv1495 or its mutant genes on the growth of M. smegmatis mc2 155 (28). Wild-type or mutant Rv1495 genes were cloned into pMind (29) to produce corresponding recombinant plasmids which were then transformed into M. smegmatis. The strain containing the empty pMind plasmid was used as negative control. For all assays, tetracycline was used to induce gene expression (30). The growth of the recombinant mycobacterial strains was examined in the presence (induction) or absence (no induction) of tetracycline (Tc). Cells were grown at 37°C with aeration in 7H9-Kan-Tw (7H9 medium supplemented with 0.5% Tween 80, 30 μg/ml kanamycin and 0.2% glycerol) to an OD600 of 1.5–2.0. Cells were diluted in 7H9 medium to an OD600 of 0.2 and grown at 37°C at 200 rpm for a further 2 h. Aliquots were taken at the indicated times, and the OD600 was measured. Each analysis was performed in triplicate. Representative growth curves were plotted.
Co-IP assays
The in vivo interactions between Rv1495 and MsmTopA were analyzed by Co-IP according to previously published procedures with some modifications (20). Exponentially growing cells of M. smegmatis with recombinant plasmid pMind-Rv1495 were harvested, resuspended and lysed in 4 ml of buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40]. Co-IPs were performed by incubating and shaking 10 μg of mycobacterial cell extract with 3 μl of MsmTopA antiserum in 100 μl buffer for 3 h at 4°C. A 20-μl slurry of protein A Sepharose was added, and incubation was continued for another hour. Immune complexes were collected, and the beads were washed with the same buffer. Finally, the beads were resuspended in SDS–PAGE sample buffer. After boiling, the samples were analyzed by western blotting using anti-Rv1495 antibody.
Cleavage of synthetic RNA by Rv1495
Oligoribonucleotides (11 bases in length) were commercially synthesized and 5′-labeled with [γ-32P]ATP using T4 polynucleotide kinase. Endoribonuclease activity was assayed in 10 μl of the reaction mixture containing 1–5 μl of Rv1495 and 32P-labeled oligonucleotides in 10 mM Tris–HCl (pH 7.8) (3). Reactions were carried out at 37°C for 30 min and stopped by the addition 10 μl of loading buffer (95% formamide, 20 mM EDTA, 0.05% bromphenol blue). The reaction mixtures were then subjected to 20% denaturing PAGE electrophoresis followed by autoradiography.
RESULTS
Rv1495 physically interacts with MtbTopA
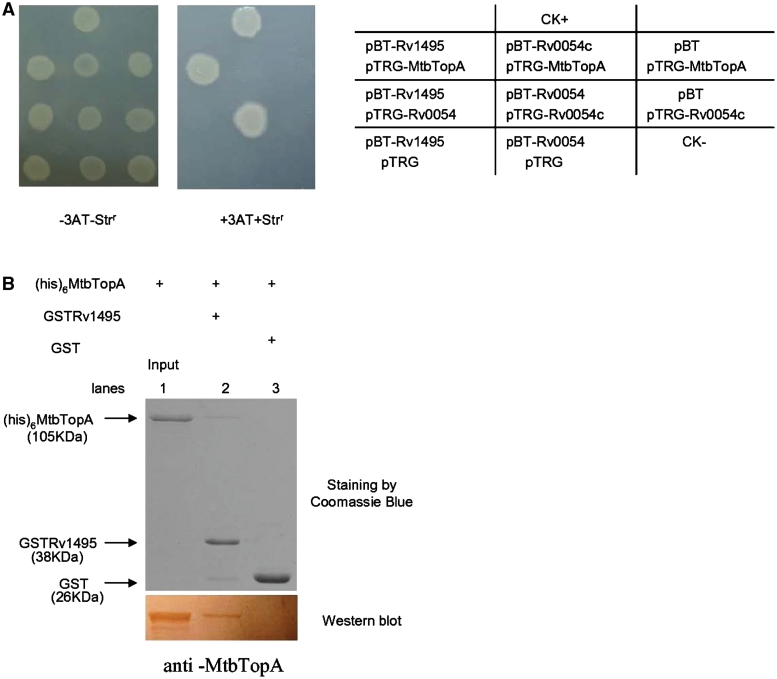
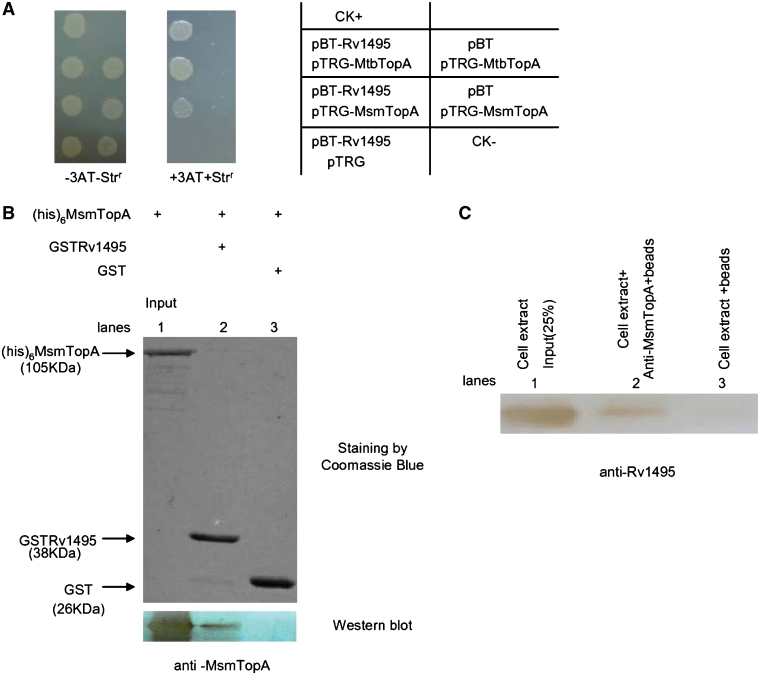
To uncover the new and as yet unknown cellular effects of the M. tuberculosis MazF genes, we conducted a screening using a bacterial two-hybrid assay to isolate proteins that interact with Rv1495, a MazF homolog. As shown in Figure 1A, the co-transformants containing Rv1495/MtbTopA grew well on the screening medium (Rv3646 encodes the topoisomerase I (MtbTopA) in the M. tuberculosis genome). Positive co-transformants (CK+) grew on the medium, whereas negative co-transformants (CK−) were incapable of growth on the same screening medium (Figure 1A). No growth was observed for their self-activated controls, or for their co-transformants expressing a non-specific protein, Rv0054, which could form polymers with itself. A clear interaction between Rv0054 with itself was detected (Figure 1A). These results show that the selective system presented here worked well and that Rv1495 was found to interact with the M. tuberculosis TopA.
Figure 1.
Physical interactions of Rv1495 with MtbTopA. (A) Bacterial two-hybrid assays (Stratagene) for the interaction of Rv1495 with MtbTopA, which were performed as described under ‘Materials and Methods’ section. Left panel, plate minus streptomycin (str) and 5 mM 3-amino-1,2,4-triazole (3-AT), Middle panel, plate plus str and 5 mM 3AT. Right panel, an outline of the plates. CK+, co-transformant containing pBT-LGF2 and pTRG-Gal11P as a positive control. CK−, co-transformant containing pBT and pTRG as a negative control. (B) Pull-down assays for examining the interaction between Rv1495 and MtbTopA. Equimolar amounts of 6× His-MtbTopA combined with GST-Rv1495 were used for pull-down assays as described in the ‘Materials and Methods’ section. GST was used as the negative control. One predicted size of his-tagged MtbTopA protein, pulled down by GST-tagged Rv1495 protein, was further examined by a western blotting assay.
Further confirmation of this phenomenon was obtained by observing direct physical interaction between Rv1495 and MtbTopA through a GST pull-down assay. As shown in Figure 1B, GST-Rv1495 and His-tagged MtbTopA proteins were co-purified. One predicted size of His-tagged MtbTopA protein could be readily pulled down by GST-Rv1495, which was further confirmed by a western blotting assay (Figure 1B, lower panel, lane 2). Co-incubation of GST with His-tagged MtbTopA did not produce any specific bands (Figure 1B, lane 3).
Rv1495 inhibits the activities of MtbTopA
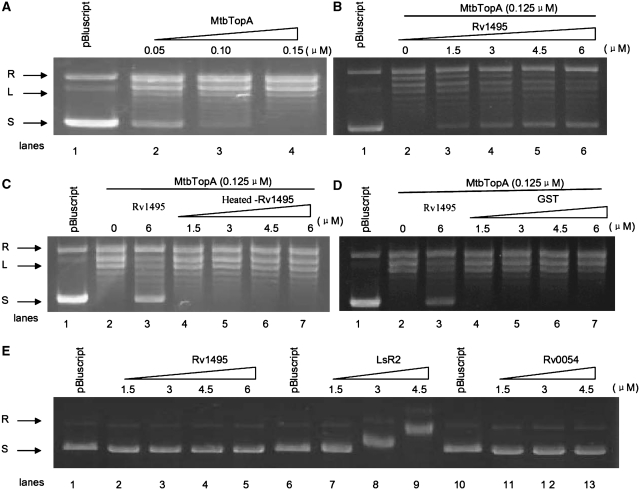
The physical interactions between Rv1495 and MtbTopA suggested that the MazF protein was in some way regulating the activities of the MtbTopA topoisomerase. As shown in Figure 2, addition of increasing amounts of MtbTopA (0.05–0.15 μM) to a reaction mixture containing plasmid pBluescript as a DNA substrate resulted in a progressive disappearance of the supercoiled plasmid DNA band, concomitant with the appearance of a relaxed plasmid band on the gel (Figure 2A). This confirmed that the MtbTopA could relax supercoiling of this DNA substrate. This activity was again examined in the presence of Rv1495 under similar conditions. As shown in Figure 2, when the concentration of MtbTopA was kept constant (0.125 μM), the amount of supercoiled plasmid steadily increased with increasing additions of Rv1495 (Figure 2B). In contrast, heat-denatured Rv1495 (Figure 2C) or GST protein (Figure 2D) had no effect on supercoiling relaxation activity of MtbTopA. The inhibitory effect was stronger if MtbTopA was preincubated with Rv1495 (Supplementary Figure S1). In further experiments, Rv1495 was found to form a protein complex with its antitoxin, Rv1494 (Supplementary Figure S2A and B), but the antitoxin could not block or reverse the effect of Rv1495 on the MtbTopA activity (Supplementary Figure S2C).
Figure 2.
Effects of Rv1495 on topoisomerase activities of MtbTopA. Topoisomerase activity assays were performed as described under ‘Materials and Methods’ section. The plasmid pBluscript was used as DNA substrate in all reaction mixtures. The reaction was treated with 6% SDS and 4 mg/ml proteinase K analyzed on a 0.8% agarose gel and then stained with ethidium bromide. Lanes 1, no topoisomerase as negative control. L, linearized plasmid; R, relaxed plasmid; S, supercoiled plasmid. (A) Different amounts of topoisomerase I (0–0.15 μM) were added to 200 ng of supercoiled pBluscript DNA. (B) Effects of different amounts of Rv1495 (0–6 μM) on the topoisomerase I (0.125 μM) activities on the supercoiled pBluscript DNA. (C) Effects of different amounts of heated-Rv1495 on the topoisomerase I activities. (D) Effects of different amounts of GST on the topoisomerase I activities. (E) EMSA assays for the plasmid DNA-binding activities of RV1495, Lsr2, or Rv0054. Lsr2 is a positive control. Rv0054 (MtbSSB) is a negative control.
A previous report showed that Lsr2 inhibited the activity of MtbTopA through its double-stranded DNA-binding ability (24). However, DNA-binding assays using pBluescript plasmid showed no similar activity for Rv1495 (Figure 2E, lanes 2–5), but confirmed this activity for Lsr2 (Figure 2E, lane 7–9). When Rv0054 was used as a negative control, no double-stranded DNA-binding activity was observed (Figure 2E, lanes 11–13).
Rv1495 inhibits single-stranded DNA cleavage by MtbTopA
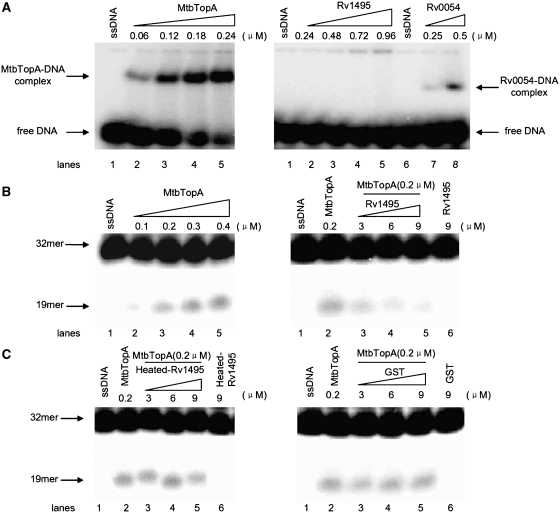
The function of the topoisomerase I depends on its binding to single-stranded DNA and subsequent cleavage activities (18,27). The mechanism underlying the inhibition of MtbTopA by Rv1495 was further analyzed by exploring the cleavage activity of MtbTopA. As shown in Figure 3A, a DNA-binding activity on the 32-mer single-stranded DNA substrate was demonstrated with a positive control, Rv0054 (MtbSSB) (Figure 3A, right panel, lanes 7–8). No DNA-binding activity was seen with Rv1495, even at a high protein concentration (0.96 μM) (Figure 3A, right panel, lane 2–5). However, a substantial shifted protein–DNA complex band, indicative of DNA-binding, was observed on the gel when an increasing concentration of MtbTopA (0.06–0.24 μM) was added to the reaction mixture (Figure 3A, left panel). This confirmed that the MtbTopA had a good DNA-binding activity with this substrate. A complicated effect was observed with Rv1495 on the DNA-binding activity of MtbTopA (Supplementary Figure S3).
Figure 3.
Effects of Rv1495 on the single-stranded DNA cleavage activity of MtbTopA. 5′-end-labeled 32-mer oligonucleotide (5′-CAGTGAGCGAGCTTCCGCTTGACATCCC AATA-3′) (27) was used as a specific single-stranded DNA substrate. DNA-binding and cleavage assays of MtbTopA were performed as described under ‘Materials and Methods’ section. (A) DNA-binding activity assays for MtbTopA, Rv1495 and Rv0054 (a positive control). The protein concentrations were indicated on top of the panels. (B) ssDNA cleavage assays for MtbtopA (left panel) and the effects of Rv1495 (right panel). The 32-mer substrates and 19-mer cleavage products were indicated by arrows. (C) The effects of the denatured Rv1495 protein (left panel) or GST (right panel) on the cleavage activity of MtbTopA.
The cleavage activity of MtbTopA was further characterized. As shown in Figure 3B (left panel), a stepwise increase in amounts of smaller DNA fragment (19-mer) was observed as 0.1–0.4 μM MtbTopA was added to the reaction mixture. When the concentration of MtbTopA was kept constant, the cleavage products steadily decreased in response to addition of increasing amounts of Rv1495 (3–9 μM) (Figure 3B, right panel), which indicated that Rv1495 inhibited the cleavage activity of MtbTopA. No effect was observed when either a heat-denatured Rv1495 protein or an unrelated protein, GST, was added (Figure 3C).
MtbTopA inhibits mRNA cleavage by Rv1495
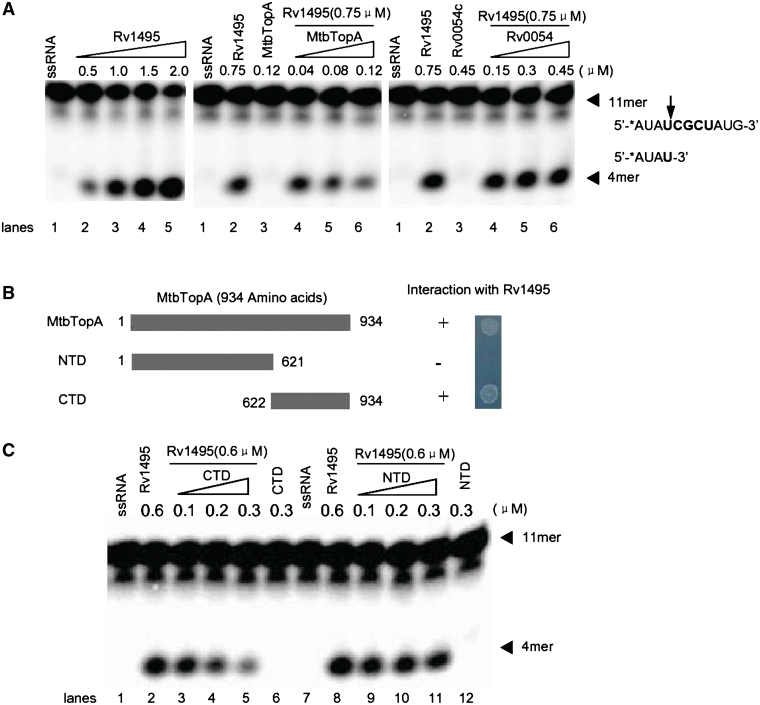
We characterized the effect of MtbTopA on the previously reported mRNA cleavage activity of Rv1495 (10). As shown in Figure 4A, when an increasing concentration of Rv1495 (0.5–2.0 μM) was added into the reaction mixture, the levels of the 11-mer mRNA substrate progressively decreased, with concomitant appearance of a band of lower molecular weight (Figure 4A, left panel, lanes 2–5), consistent with mRNA cleavage. When the concentration of Rv1495 was kept constant (0.75 μM), the level of cleavage products steadily decreased as MtbTopA content was increased (Figure 4A, middle panel, lanes 4–6), confirming that the MtbTopA inhibited the mRNA cleavage activity of Rv1495. In contrast, Rv0054, used as a negative control, had no inhibitory effect on cleavage (Figure 4A, right panel, lanes 4–6). The inhibitory domain of MtbTopA was further characterized by purifying the NTD of MtbTopA, and its CTD (Figure 4B). The interactions of these two domains with Rv1495 were then characterized by a bacterial two-hybrid assay. As shown in Figure 4B, CTD interacted with Rv1495, while NTD did not. CTD also showed an equivalent inhibitory effect on the mRNA cleavage by Rv1495 as was seen with the wild-type MtbTopA (Figure 4C, lanes 2–5). No significant effect was observed for the NTD protein under similar reaction conditions (Figure 4C, lanes 9–12).
Figure 4.
Effects of MtbTopA on mRNA cleavage activities of Rv1495. (A) mRNA cleavage experiments were performed as described under ‘Materials and Methods’ section. A variant concentration of Rv1495 (0.5–2.0 μM) or MtbTopA (40–120 nM) proteins was incubated with 0.5 μM mRNA substrates in the reactions. The mRNA substrate, products and their sequences were indicated by arrows. Rv0054 was used a negative control. (B) Two mutant proteins of MtbTopA and their interactions with Rv1495. The names given to mutants (NTD and CTD) and their amino acid regions are indicated. Bacterial two-hybrid assays for all mutants were performed as described under ‘Materials and Methods’ section. + represents growth on the screening plate; − represents no growth on the screening plate. (C) Effects of N-terminal and C-terminal mutant proteins of MtbTopA on the mRNA cleavage activities of Rv1495. The protein concentrations were indicated on top of the panel.
Therefore, the mRNA cleavage activity of Rv1495 was inhibited through the CTD of MtbTopA.
Rv1495 interacts with M. smegmatis topoisomerase I (MsmTopA) both in vitro and in vivo
Mycobacterium smegmatis also contains an ortholog of the M. tuberculosis H37Rv topoisomerase I, MsmTopA (17). To detect if the interaction is conserved in both mycobacterial strains, we examined the interaction between Rv1495 with the M. smegmatis MsmTopA using a bacterial two-hybrid assay. As shown in Figure 5A, the co-transformant with Rv1495 and MsmTopA genes obtained a similar growth on the selective medium to that seen with the Rv1495 and MtbTopA cotransformant. No growth was observed for several negative controls (Figure 5A). This indicated that M. tuberculosis Rv1495 might interact with MsmTopA. Further confirmation of the conserved interaction was obtained by conducting a GST pull-down assay. As shown in Figure 5B, GST-Rv1495 and His-tagged MsmTopA proteins were co-purified. A protein of the predicted size of the His-tagged MsmTopA protein could be readily pulled down by GST-Rv1495, which was confirmed by a further western blotting assay (Figure 5B lower panel, lane 2). GST co-incubated with his-tagged MtbTopA did not produce any specific bands (Figure 5B, lane 3).
Figure 5.
Interactions of Rv1495 with MsmTopA in vitro and in vivo. (A) Bacterial two-hybrid assays (Stratagene). Experiments were performed as described under ‘Materials and Methods’ section. Left panel, plate minus streptomycin (str) and 5 mM 3-amino-1,2,4-triazole (3-AT), Middle panel, plate plus str and 5 mM 3AT. Right panel, an outline of the plates. (B) Pull-down assays for examining the interaction between Rv1495 and MsmTopA. Equimolar amounts of 6× His-MsmTopA combined with GST-Rv1495 were used for pull-down assays as described in the ‘Materials and Methods’ section. GST was used as the negative control. One predicted size of his-tagged MsmTopA protein, pulled down by GST-tagged Rv1495 protein, was further examined by a western blotting assay. (C) Co-IP assays. Exponentially growing cells of the recombinant M. smegmatis containing Rv1495-expression plasmid were harvested, resuspended and lysed. Co-IPs were performed by incubating 10 μl of M. smegmatis cell extracts with 3 μl of MsmTopA antiserum for 3 h at 4°C. A 20-μl slurry of protein A Sepharose was added, and incubation was continued for another hour. Immune complexes were collected, and the beads were washed with buffer. Finally, the beads were resuspended in SDS–PAGE sample buffer. After boiling, the samples were analyzed by western blotting using anti-Rv1495 antibody.
For further investigation of the physiological significance of these in vitro reactions, we examined co-immunoprecipitation of the MsmTopA with Rv1495, which was expressed through a pMind recombinant plasmid in M. smegmatis (29). As shown in Figure 5C, the specific hybridization signal for the expressed Rv1495 in M. smegmatis cell extracts was detected by anti-Rv1495 antibody (lane 1). Using Protein A beads that were first conjugated with antibody raised against MsmTopA, an in vivo physical interaction between MsmTopA and Rv1495 was tested. MsmTopA was shown to associate with Rv1495 in vivo because an obvious and a specific hybridization signal was detected (Figure 5C, lane 2), although to a lesser extent than the signal for the expressed Rv1495 in M. smegmatis cell extracts (Figure 5C, lane 1). In contrast, no obvious specific signal was detected for the association if the anti-MsmTopA antibody was absent from the reactions (Figure 5C, lane 3). Therefore, M. tuberculosis Rv1495 could interact with the homolog of M. smegmatis topoisomerase I both in vitro and in vivo.
Characterization of an inhibitory peptide fragment from Rv1495
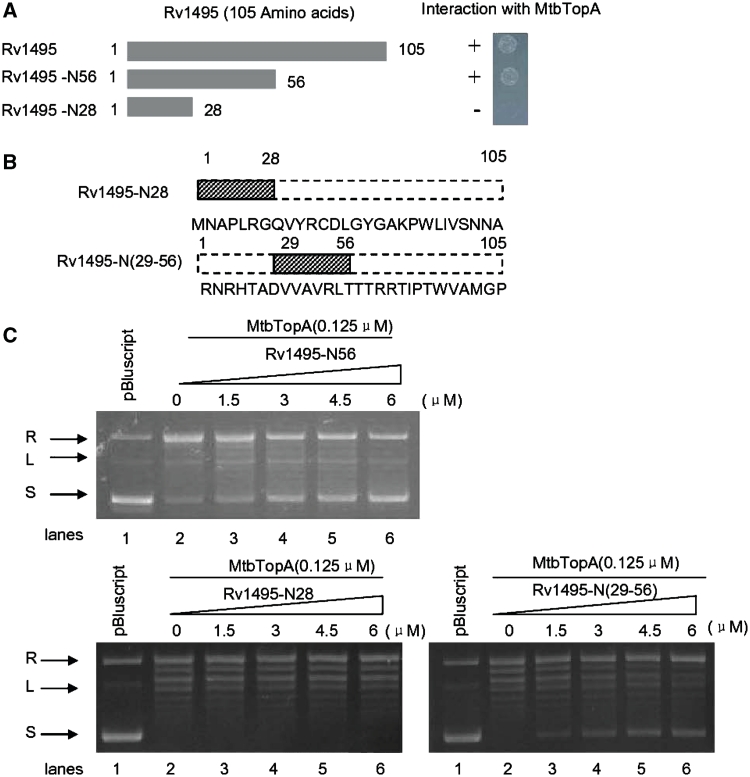
To map the interactive domain of Rv1495 with MtbTopA, we produced several mutant fragments of the Rv1495 gene (Figure 6A). Using bacterial two-hybrid assays, we found that the N-terminal 56 amino acid fragment of Rv1495 interacted with MtbTopA (Figure 6A, right panel). We designed and synthesized two shorter peptides, Rv1495-N28 and Rv1495-N(29-56), as shown in Figure 6B, and tested these for inhibitory action on MtbTopA activity. Rv1495-N(29-56) retained an inhibitory effect on the topoisomerase I activity (Figure 6C, lower right panel).
Figure 6.
N-terminus of Rv1495 regulates the topoisomerase activities of MtbTopA. Topoisomerase activity and bacterial two-hybrid assays for all mutants were performed as described under ‘Materials and Methods’ section. + represents growth on the screening plate; − represents no growth on the screening plate. (A) The N-terminal mutant fragments of Rv1495 containing 56 and 28 amino acid residues (left panel) and their interactions with MtbTopA assayed by the bacterial two-hybrid system (right panel). (B) Two synthesized short peptides and their amino acid sequences. (C) Effect of different amount of Rv1495-N28, Rv1495-N56, and Rv1495-N(29-56) (0–6.0 μM) on topoisomerase activity.
Rv1495-N(29-56) loses the mRNA cleavage activity but retains its ability to inhibit MsmTopA activity and mycobacterial growth
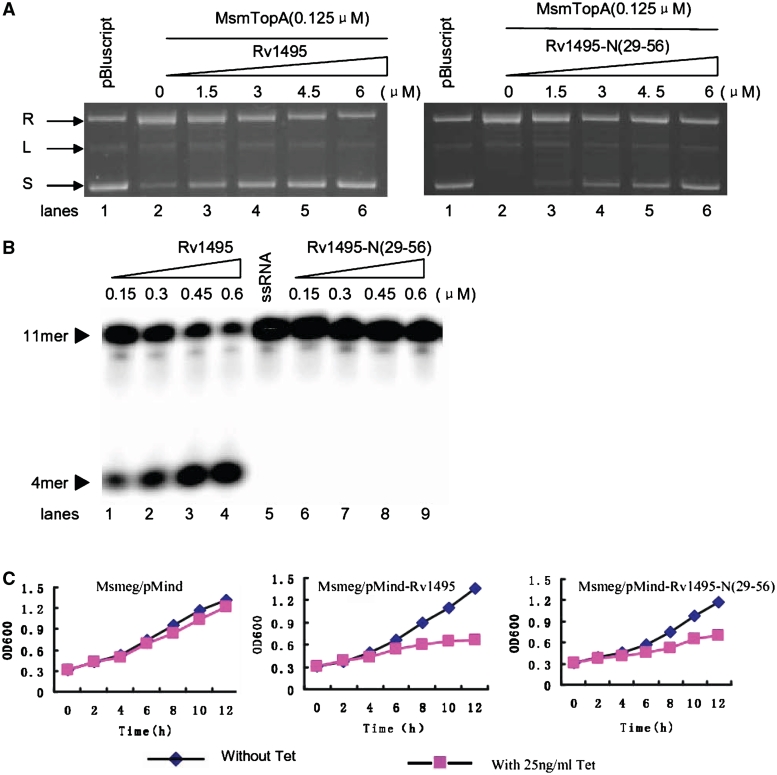
As shown in Figure 6, Rv1495-N(29-56) retained its ability to interact with MtbTopA and to inhibit topoisomerase activity. We compared the effects of two proteins, Rv1495 and Rv1495-N(29-56), on the topoisomerase activity of MsmTopA. As shown in Figure 7A, the activity of MsmTopA was progressively inhibited as the Rv1495 concentration was increased, as indicated by the appearance of increasing amounts of supercoiled DNA. Similarly, an inhibition on the function of MsmTopA was also observed in the presence of Rv1495-N(29-56) (Figure 7A, right panel).
Figure 7.
Activities of Rv1494 and Rv1495-N(29-56) and their effects on the mycobacterial growth. (A) Effects of Rv1495 and Rv1495-N(29-56) on the topoisomerase activities of MsmTopA. The assays were performed as described under ‘Materials and Methods’ section. L, linearized plasmid; R, relaxed plasmid; S, supercoiled plasmid. The protein concentrations were indicated on top of the panels. (B) mRNA cleavage activity assays. Experiments were performed as described under ‘Materials and Methods’ section. A varying concentration of Rv1495 or Rv1495-N(29-56) proteins was incubated with 0.5 μM mRNA substrates in the reactions. The mRNA substrate and products were indicated by arrows. (C) Effects of the expressions of Rv1495 and Rv1495-N(29-56) on the mycobacterial growth. A TetR-controlled expression system was used to analyze the effects of Rv1495 genes on the growth of M. smegmatis mc2 155 as described in the ‘Materials and Methods’ section. The growth of these recombinant mycobacterial strains were examined in the presence (induction) or absence (no induction) of tetracycline (Tc). Aliquots were taken at the indicated times and the OD600 was measured. Each analysis was performed in triplicate. The representative growth curves were plotted. The recombinant mycobacterial strains were indicated above the panels.
A further activity assay indicated that Rv1495-N(29-56) lost its mRNA cleavage ability (Figure 7B, lanes 5–9). In contrast, the wild-type protein (Rv1495) demonstrated good activity as increasingly shorter mRNA excision products were observed on the gel as the Rv1495 concentrations were increased stepwise (Figure 7B, lanes 1–4). Addition of Rv1495 also had clear effects on the mycobacterial growth, as indicated by an obvious inhibition following induction of Rv1495 expression by tetracycline in M. smegmatis through the pMind recombinant plasmid (Figure 7C, middle panel). An important observation was that Rv1495-N(29-56), although lacking mRNA cleavage activity, was still clearly able to inhibit the growth of M. smegmatis (Figure 7C, right panel). This growth inhibition could be rescued by coexpression of the C-terminus of MtbTopA (Supplementary Figure S4). In contrast, the recombinant strain with the pMind alone showed only a slight effect on growth (Figure 7C, left panel). In further experiments, no significant effect was observed on the growth of E. coli following induction of Rv1495 expression (Supplementary Figure S5). This lack of effect is most likely because there is no homology between the C-terminus of Mycobacterium and the E. coli TopA enzymes. Therefore, Rv1495 or its N-terminal truncation could negatively regulate the topoisomerase activity of MsmTopA and inhibit mycobacterial growth.
DISCUSSION
In this study, we have shown that a MazF homolog, Rv1495, from M. tuberculosis could physically interact with TopA and that this interaction resulted in mutual inhibition of the respective activities of both proteins. Rv1495 physically interacted with MsmTopA, both in vitro and in vivo, and inhibited mycobacterial growth when expressed in M. smegmatis. An N-terminal fragment of Rv1495 was further characterized and shown to retain the capacity for both physical interaction with, and inhibition of, TopA, even though it had lost its mRNA cleavage activity. Therefore, in addition to its known RNA interferase function, the bacterial MazF protein also can directly interact with and modulate the activities of topoisomerase I. This suggests an additional regulatory function of the M. tuberculosis MazEF system on bacterial growth that functions separately from its ribonuclease activity.
The MazEF system may be involved in ability of M. tuberculosis to undergo long-term dormancy in human tissues, which is essential for the persistence of the pathogen (31,32). The activity of the sole TopA in this species with respect to relaxation of negative supercoiling might be important during stress responses, since a number of stress genes have to be induced for survival (3,33). In the current study, we found that the function of Rv1495, a toxin protein, was linked to the topoisomerase I of the pathogen. Mycobacterium tuberculosis might have evolved a mechanism for survival within its host macrophage cells that operates by perfectly balancing the interactions of a pair of MazEF and the interactions of MazEF with the DNA topoisomerase I.
This mechanism would allow the organism to become dormant within the hostile environment of human macrophage cells. For example, MazE and MazF would normally form a stable complex, which would prevent RNA cleavage by MazF. When the pathogen confronts the stressful environment within human cells, MazE would be degraded by the ATP-dependent ClpPA serine protease (1). This would result in the release of MazF from the MazE–MazF protein complex. As shown earlier (3,16), MazF, on the one hand, could cleave mRNA and inhibit gene expression. On the other hand, as shown in the current study, MazF could inhibit the activity of topoisomerase I through a direct protein–protein interaction, again inhibiting gene expression. These two mechanisms would then contribute to pathogen dormancy.
The importance of TA systems in the life cycle of M. tuberculosis is suggested by the presence of the remarkably large number of these genes (15). In contrast, despite a much larger genome (7 versus 4.4 Mb) M. smegmatis has only two TA systems (2,34), including one MazF homolog. However, although M. tuberculosis and M. smegmatis have multiple groups of MazEF systems, both of their genomes encode a single highly homologous type I topoisomerase. Interestingly, in the current study, we found that Rv1495 interaction was conserved between both MtbTopA and MsmTopA and that it had a similar inhibitory action on topoisomerase activity of both organisms. An in vivo interaction between Rv1495 and the MsmTopA of M. smegmatis was also demonstrated. Thus, this interaction between these two proteins might have physiological importance in M. tuberculosis.
Mycobacterium smegmatis has been used as a suitable host for the TA growth assays since it contains only two TA systems (2,34). In the current study, we observed that Rv1495 could inhibit the growth of M. smegmatis when Rv1495 was expressed through a pMind recombinant plasmid. Importantly, an N-terminal fragment of Rv1495, Rv1495-N(29-56), was also effective at inhibiting mycobacterial growth (Figure 7C), although it was less effective than the full-length Rv1495 (Figure 7A). Since Rv1495-N(29-56) had lost its mRNA cleavage activity, growth inhibition is most likely attributable to the effects of the MazF protein on the functioning of the topoisomerase (Figure 7B). Therefore, two different mechanisms would appear to contribute to the inhibition of mycobacterial growth by Rv1495. On the one hand, its interferase activity would cleave mRNA. On the other hand, its physical interaction with TopA would negatively regulate the relaxation of supercoiled DNA carried out by the topoisomerase.
The incidence of tuberculosis has been increasing substantially on a worldwide basis over the past decade. To complicate matters, drug-resistant TB has also become a major public health concern. However, no new tuberculosis-specific drugs have been discovered for more than 40 years. Bacterial topoisomerase I is required for preventing hypernegative supercoiling of DNA during transcription (35), which has been considered to be especially important for the pathogenic adaptation to stressful environments (33,36). The genome of M. tuberculosis encodes a type I topoisomerase, but all attempts to isolate a transposon insertion mutant in the topA of M. tuberculosis gene have been unsuccessful (20). This suggests that M. tuberculosis topoisomerase I is critical for survival. Recent studies have found that interference with the function of topoisomerase I and stabilization of its covalent complex with cleaved DNA can lead to bacterial cell death (37–39). Thus, topoisomerase I of pathogens is now becoming recognized as a promising new target for combating bacterial drug-resistance. In the current study, we characterized a N-terminal fragment, Rv1495-N(29-56), which retained the interaction with TopA and the inhibitory activity on the topoisomerase function. These specific protein–protein and protein–peptide interactions provided us with important clues for developing effective antibiotics that could target TB topoisomerase I. Compounds that activate or mimic bacterial toxins could quite feasibly be developed into novel antibiotics. On the one hand, drugs of this type would promote pathogenic programmed cell death by preventing or reducing the association between a given MazEF pair. On the other hand, they would also promote an inhibition of the function of the topoisomerase I and thus interfere with transcriptional regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China (30930003); 973 Program (2006CB504402); National Special Key Project of China on Major Infectious Diseases (2008ZX10003-005). Funding for open access charge: National Special Key Project of China on Major Infectious Diseases (2008ZX10003-005).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′, 5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl Acad. Sci. USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu L, Zhang Y, Teh JS, Zhang J, Connell N, Rubin H, Inouye M. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 2006;281:18638–18643. doi: 10.1074/jbc.M512693200. [DOI] [PubMed] [Google Scholar]
- 4.Engelberg-Kulka H, Hazan R, Amitai S. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. Cell Sci. 2005;118:4327–4332. doi: 10.1242/jcs.02619. [DOI] [PubMed] [Google Scholar]
- 5.Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 6.Tsilibaris V, Maenhaut-Michel G, Mine N, Van Melderen L. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J. Bacteriol. 2007;189:6101–6108. doi: 10.1128/JB.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marianovsky I, Aizenman E, Engelberg-Kulka H, Glaser G. The regulation of the Escherichia coli mazEF promoter involves an unusual alternating palindrome. J. Biol. Chem. 2001;276:5975–5984. doi: 10.1074/jbc.M008832200. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Zhang Y, Inouye M. Characterization of the interactions within the mazEF addiction module of Escherichia coli. J. Biol. Chem. 2003;278:32300–32306. doi: 10.1074/jbc.M304767200. [DOI] [PubMed] [Google Scholar]
- 9.Kamada K, Hanaoka F, Burley SK. Crystal structure of the MazE/MazF complex:molecular bases of antidote-toxin recognition. Mol. Cell. 2003;11:875–884. doi: 10.1016/s1097-2765(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 10.Zhu L, Phadtare S, Nariya H, Ouyang M, Husson RN, Inouye M. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 2008;69:559–569. doi: 10.1111/j.1365-2958.2008.06284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 12.James PE, Grinberg OY, Michaels G, Swartz HM. Intraphagosomal oxygen in stimulated macrophages. J. Cell Physiol. 1995;163:241–247. doi: 10.1002/jcp.1041630204. [DOI] [PubMed] [Google Scholar]
- 13.Schaible UE, Sturgill-Koszycki S, Schlesinger PH, Russell DG. Cytokine activation leads to acidification and increases maturation of Mycobacterium avium-containing phagosomes in murine macrophages. J. Immunol. 1998;160:1290–1296. [PubMed] [Google Scholar]
- 14.Via LE, Fratti RA, McFalone M, Pagan-Ramos E, Deretic D, Deretic V. Effects of cytokines on mycobacterial phagosome maturation. J. Cell Sci. 1998;111:897–905. doi: 10.1242/jcs.111.7.897. [DOI] [PubMed] [Google Scholar]
- 15.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier C, Gas S, Barry CE, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 16.Zhao L, Zhang J. Biochemical characterization of a chromosomal toxin–antitoxin system in Mycobacterium tuberculosis. FEBS Lett. 2008;582:710–714. doi: 10.1016/j.febslet.2008.01.045. [DOI] [PubMed] [Google Scholar]
- 17.Bhaduri T, Bagui TK, Sikder D, Nagaraja V. DNA topoisomerase 1 from Mycobacterium smegmatis. An enzyme with distinct features. J. Biol. Chem. 1998;273:13925–13932. doi: 10.1074/jbc.273.22.13925. [DOI] [PubMed] [Google Scholar]
- 18.Jain P, Nagaraja V. Indispensable, functionally complementing N and C-terminal domains constitute site-specific topoisomerase I. J. Mol. Biol. 2006;357:1409–1421. doi: 10.1016/j.jmb.2006.01.079. [DOI] [PubMed] [Google Scholar]
- 19.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc. Natl Acad. Sci. USA. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Zhang L, Liu Y, Yang S, Gao C, Gong H, Feng Y, He ZG. Archaeal eukaryote-like Orc1/Cdc6 initiators physically interact with DNA polymerase B1 and regulate its functions. Proc. Natl Acad. Sci. USA. 2009;106:7792–7797. doi: 10.1073/pnas.0813056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo M, Feng H, Zhang J, Wang W, Li Y, Gao C, Chen H, Feng Y, He ZG. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009;19:1301–1308. doi: 10.1101/gr.086595.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui T, Zhang L, Wang X, He ZG. Uncovering new signaling proteins and potential drug targets through the interactome analysis of Mycobacterium tuberculosis. BMC Genomics. 2009;10:118. doi: 10.1186/1471-2164-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 25.Colangeli R, Helb D, Vilchèze C, Hazbón MH, Lee CG, Safi H, Sayers B, Sardone I, Jones MB, Fleischmann RD. Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis. PLoS Pathog. 2007;3:e87. doi: 10.1371/journal.ppat.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grasser KD, Ritt C, Krieg M, Fernandez S, Alonso JC, Grimm R. The recombinant product of the Chryptomonas phi plastid gene hlpA is an architectural HU-like protein that promotes the assembly of complex nucleoprotein structures. Eur. J. Biochem. 1997;249:70–76. doi: 10.1111/j.1432-1033.1997.00070.x. [DOI] [PubMed] [Google Scholar]
- 27.Bhat AG, Leelaram MN, Hegde SM, Nagaraja V. Deciphering the distinct role for the metal coordination motif in the catalytic activity of Mycobacterium smegmatis topoisomerase I. J. Mol. Biol. 2009;393:788–802. doi: 10.1016/j.jmb.2009.08.064. [DOI] [PubMed] [Google Scholar]
- 28.Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, Schnappinger D. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blokpoel MC, Murphy HN, O'Toole R, Wiles S, Runn ES, Stewart GR, Young DB, Robertson BD. Tetracycline-inducible gene regulation in mycobacteria. Nucleic Acids Res. 2005;33:e22. doi: 10.1093/nar/gni023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gossen M, Bujard H. Anhydrotetracycline, a novel effector for tetracycline controlled gene expression systems in eukaryotic cells. Nucleic Acids Res. 1993;21:4411–4412. doi: 10.1093/nar/21.18.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 32.Spoering AL, Lewis K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 2001;183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tse-Dinh YC. Bacterial topoisomerase I as a target for discovery of antibacterial compounds. Nucleic Acids Res. 2009;37:731–737. doi: 10.1093/nar/gkn936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korch SB, Contreras H, Clark-Curtiss JE. Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J. Bacteriol. 2009;191:1618–1630. doi: 10.1128/JB.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruss GJ, Drlica K. Topoisomerase I mutants: the gene on pBR322 that encodes resistance to tetracycline affects plasmid DNA supercoiling. Proc. Natl Acad. Sci. USA. 1986;83:8952–8956. doi: 10.1073/pnas.83.23.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang JC. DNA topoisomerases. Annu. Rev, Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 37.Tse-Dinh YC. Exploring DNA topoisomerases as targets of novel therapeutic agents in the treatment of infectious diseases. Infect. Disord. Drug Targets. 2007;7:3–9. doi: 10.2174/187152607780090748. [DOI] [PubMed] [Google Scholar]
- 38.Baaklini I, Usongo V, Nolent F, Sanscartier P, Hraiky C, Drlica K, Drolet M. Hypernegative supercoiling inhibits growth by causing RNA degradation. J. Bacteriol. 2008;190:7346–7356. doi: 10.1128/JB.00680-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 2006;59:723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.