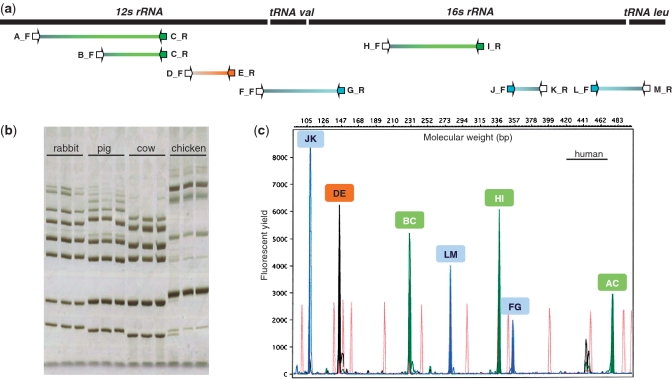
Figure 5.
Experimental application and validation of a SPInDel profiling kit for identification of eutherian species. (a) Graphical representation of the seven ribosomal RNA hypervariable regions amplified by multiplex PCR. PCR primers (arrows) were named using letters A to M, their orientation (F, for forward, R for reverse) and labeling with fluorescent dyes (blue, orange and green arrows). (b) The products of multiplex PCRs of three eutherian and one avian species are shown on a silver stained polyacrylamide gel. Each species has a unique pattern of migration because of differences in the length of amplicons (hypervariable regions). Identification of species using such gels is only possible by comparing the banding pattern of the target sample with those of reference samples analyzed with the same procedure. (c) Electropherogram illustrating a SPInDel profile from a human reference sample obtained by capillary electrophoresis with multidye fluorescence detection. The profile is displayed in a four-color fluorescent system, in which green, blue and yellow channels were used for detection of amplified products and red was used for a size marker. The species identification is achieved by running the SPInDel numeric profile of the target sample against a reference database (SPInDel workbench).