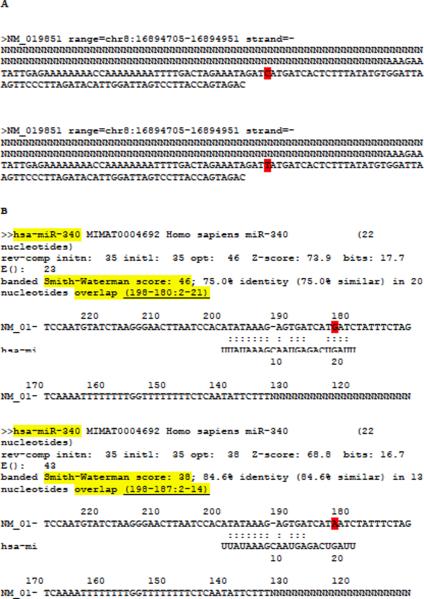
Figure 5.
MicroSNiPer pre-processing of FGF20 input sequence before running with FASTA program and its FASTA output. (A) Two FGF20 input sequences in separate files generated with SNP rs12720208 in position 182[C/T]. (B) Two FASTA program output files corresponding to each input file, respectively. The 3'UTR sequence and the alternative alleles are complementary to the input 3'UTR sequence. The FGF20 3'UTR sequence is masked upstream with `N'. The alternative alleles are highlighted in red. MicroSNiPer builds a unique lookup key (highlighted with light yellow background) composed of the miRNA name, the banded Smith-Waterman score, and the coordinates of the overlap.