Abstract
Vaccination is one of the greatest triumphs of modern medicine, yet we remain largely ignorant of the mechanisms by which successful vaccines stimulate protective immunity. Two recent advances are beginning to illuminate such mechanisms: realization of the pivotal role of the innate immune system in sensing microbes and stimulating adaptive immunity, and advances in systems biology. Recent studies have used systems biology approaches to obtain a global picture of the immune responses to vaccination in humans. This has enabled the identification of early innate signatures that predict the immunogenicity of vaccines, and identification of potentially novel mechanisms of immune regulation. Here we review these advances, and critically examine the potential opportunities and challenges posed by systems biology in vaccine development.
“We are drowning in a sea of data and thirsting for knowledge. Most biology today is low input, high throughput, no output biology.” Sydney Brenner
“We must make this the decade of vaccines.” Bill Gates
Introduction
In the epic saga of the evolutionary struggle between microbes and humans, the invention of vaccination is a defining moment, one that represents the victory of our wits over their genes. Ironically however, despite the common origins of vaccinology and immunology, in the pioneering work of giants such as Pasteur and Jenner, the two disciplines have evolved such different trajectories that immunologists remain largely ignorant about the mechanisms of action of successful vaccines (empirically made), and vaccinologists have until recently, displayed little interest in the intricacies of immune regulation. Understanding the immunological mechanisms of vaccination, however, is of paramount importance in the rational design of future vaccines against pandemics such as HIV, malaria, tuberculosis and against emerging infections. Recent advances in our understanding of the innate immune system and the use of systems biological approaches are beginning to reveal the fundamental mechanisms by which the innate immune system orchestrates protective immune responses to vaccination (Pulendran and Ahmed, 2006; Steinman, 2008). The innate immune system is capable of sensing viruses, bacteria, parasites, and fungi through the expression of so-called pattern recognition receptors (PRRs), which are expressed by dendritic cells (DCs) and other cells of the innate immune system (Reviewed by Coffman and Seder – this volume of Immunity). Toll-like receptors (TLRs) represent the most studied family of PRRs (Iwasaki and Medzhitov, 2010; Kawai and Akira, 2010). However, other non-TLR families of innate receptors, such as C-type lectin-like receptors (Geijtenbeek and Gringhuis, 2009), NOD-like receptors (Ting et al., 2010), and RIG-I-like receptors (Wilkins and Gale, 2010), also play critical roles in innate sensing of pathogens and induction of inflammatory responses. Emerging evidence suggests that the nature of the DC subtype, as well as the particular PRR triggered, play critical roles in modulating the strength, quality, and persistence of adaptive immune responses (Pulendran and Ahmed, 2006; Steinman, 2008). Such insights about the molecular basis of immune regulation have accrued largely through the traditional scientific method of hypothesis creation, and experimental validation, particularly through the reductionist approaches of molecular biology. However, as powerful as such approaches are, they offer a very limited view of complex biological systems. Thus, there are estimated to be more than 26,000 genes in our genomes, and entry of a vaccine or a pathogen into the body, perturbs the expression of a substantial fraction of them. Systems biological tools offer us a solution to this problem. In vaccinology, recent studies have highlighted the use of such approaches in offering a global picture of the biological response to a vaccine. Here we highlight these advances and discuss their potential importance. This review is divided into 4 parts. In the first part (“Biology of the 21st century”), we provide a broad overview of systems biology, its goals and challenges, and highlight the features that distinguish it from reductionistic biology. Next, (in “Systems biology in vaccinology”) we review recent studies that have applied systems biological approaches to vaccinology, and suggest key areas where such approaches may impinge on vaccine development. These include identification of potentially novel correlates of immunity, predicting the efficacy of vaccines, accelerating the clinical trial platform of vaccines, and learning new biological insights about immune regulation. In part three (“Low Input, High Throughput, No Output Biology”), we critically examine the challenges and potential pitfalls of systems biological approaches. Finally (in “A framework for systems vaccinology”), we conclude by offering a conceptual framework of how systems approaches can guide vaccine design and development.
Biology of the 21st Century
Two of the greatest scientific achievements of the 20th century were the discovery of the structure of DNA, and the sequencing of the human genome. The grand challenge for biology and medicine at the turn of the 21st century is to understand the biological complexity that emerges from interactions between our genomes and the environments. We are uniquely poised to tackle this challenge of biological complexity by the convergence of a new intellectual framework (a “systems,” rather than a reductionistic view), and new technologies (for measuring and visualizing the behavior of genes, molecules, cells, organs and organisms), coupled with the innovation of computational and mathematical tools for dealing with complex data sets. The convergence of these disparate threads offers us an unprecedented opportunity to understand the fundamental features of life, from a “holistic” rather than solely reductionistic; from a predictive, rather than descriptive; in short – from a systems biological viewpoint.
Systems Biology is an interdisciplinary approach that systematically describes the complex interactions between all the parts in a biological system, with a view to elucidating new biological rules capable of predicting the behavior of the biological system (Kitano, 2002). While reductionist molecular biology works by isolating and characterizing each component of the system (e.g. a gene/protein or a cell type), systems biology focus in studying the structure and dynamics of the whole system (Kitano, 2002). Under different types of perturbation, data are collected from all the components of a biological system, analyzed and integrated in order to generate a mathematical model that describes or predicts the response of the system to individual perturbations (Ideker et al., 2001). A key goal of systems biology is to understand the nature of biological networks, which access, integrate and communicate information from the genome to the environment, and back (Ideker et al., 2001; Kitano, 2002). These networks represent, in a sense, the lowest functional units of life processes such as development, disease, immunity and aging. Therefore, understanding these life processes requires understanding the nature and behavior of these networks, both their robustness and plasticity, in the face of a dynamic environment. What is needed to delineate these networks is the acquisition of high throughput data on the genes, mRNAs, microRNAs, proteins that constitute the networks. Systems biology capitalizes on several so-called “omic” technologies which are used to define and monitor all the components of the systems. DNA microarrays and high throughput sequencing can be applied to identify global differences on gene expression (transcriptomics), genomic rearrangements and genetic polymorphisms (genomics) as well as to provide a high resolution global map of protein-DNA interactions (chromatin immunoprecipitation followed by DNA sequencing or hybridization to the array). Other enabling technologies include modern mass spectrometry (powering proteomics, lipidomics and metabolomics), yeast two-hybrid system (mapping protein interactions ) and genome-wide RNA interference screening (identifying genes required for a process). In addition, systems biology features the integration and modeling the huge amount of data generated by high-throughput techniques. An array of computational methods has been developed in the context of systems biology, of special interests data integration and network inference (Bansal et al., 2007; Hyduke and Palsson, 2010). Such methods can be closely coupled with experimental studies, to generate testable hypotheses and improve the understanding of molecular mechanisms.
Systems biological approaches have changed prognosis and therapy response prediction in oncology (Alizadeh et al., 2000; Potti et al., 2006; Sorlie et al., 2001), and are beginning to be applied to understanding mechanisms of innate and adaptive immunity (Aderem and Hood, 2001; Germain, 2001; Gilchrist et al., 2006; Haining et al., 2008; Haining and Wherry, 2010; Kaech et al., 2002; Wherry et al., 2007; Zak and Aderem, 2009), in identifying diagnostic biomarkers of different infections (Chaussabel et al., 2008; Lee et al., 2008; Otaegui et al., 2009; Ramilo et al., 2007), and autoimmunity (Pascual et al., 2010). Systems biological approaches also offer unprecedented opportunities to study immune responses in humans (Aderem and Hood, 2001; Germain, 2001). However, only recently have they started to be applied to vaccinology. There are two broad applications of systems approaches in vaccinology: prediction of immunogenicity and efficacy of vaccines, and scientific discovery. These two areas use distinct methodologies, and have different rationales and output, and are discussed below.
Systems biology in vaccinology
One potential application of systems biology in vaccinology is in predicting vaccine efficacy. The identification of molecular signatures (e.g. patterns of gene expression induced after vaccination), induced rapidly in the blood after vaccination that correlate with and predict the later development of protective immune responses, represents a strategy to prospectively determine vaccine efficacy. In the field of cancer genomics, predictions of cancer outcome have been based on gene expression profiles of the cancer cells themselves [see Box]. However in the human immune system, there is no analogous single tissue from which to sample cells for dissecting biology and creating predictors. The immune system spans multiple lineages, is anatomically distributed and is highly inter-regulated. Sampling all these cellular components and assaying their gene expression profiles is obviously not feasible. However, two critical features of the immune response provide the rationale for applying genomic approaches to study the response to vaccines. First, cells of the immune system are easily accessible in peripheral blood samples. Each blood sample provides a snap-shot of many lineages and dozens of differentiation states within the immune system. Moreover, because migration and trafficking is central and ongoing feature of the immune response, peripheral blood leukocytes represent recent emigrants of peripheral tissues, including vaccine sites. Second, cells of the immune system are uniquely sensitive to perturbation. As discussed below, we (Querec et al., 2009) and others (Gaucher et al., 2008) have demonstrated, individuals who have been vaccinated manifest marked and characteristic changes in the gene expression profiles of their peripheral blood leukocytes. Thus the population of immune cells in the peripheral blood provides a sensitive bellwether of localized or systemic immunologic events.
Box: Prediction and classification based on gene expression signatures.
In cancer genomics, gene expression signatures have been used to predict the patient’s clinical outcome and response to therapies. In vaccinology, patterns of gene expression induced after vaccination (i.e. “signatures”) could be used to predict immunogenicity or efficacy. For example, a particular gene expression signature induced early after vaccination, may be able to accurately classify vaccinees into distinct groups, e.g. “high” versus “low” responders, based on whether the antibody titers are above and below the threshold necessary to confer protective immunity. For many vaccines, such “correlates of protection,” have been established (e.g. Table 1). Such a signature can then be used to predict, in an independent trial, whether for example, a vaccinee would generate an antibody response above the threshold necessary for protection. Towards this end, we have used a machine learning approach to identify signatures that were capable of predicting the immunogenicity of individuals vaccinated with the yellow fever vaccine-17D (Querec et al, 2008) or the inactivated influenza vaccine.
The identification of signatures that predict the immunogenicity of vaccines could have broad public health utility in several situations: 1. Identification of individuals who respond sub-optimally to vaccination (e.g. elderly, infants, immune compromised); 2. Rapid screening of first responders during emergency outbreaks to identify vaccinees who respond sub-optimally; 3. Identification of non responders in partially effective vaccines (e.g. RTS malaria vaccine); 4. Accelerated assessment of vaccine immunogenicity and of efficacy (e.g. new meningococcal vaccine); 5. Identification of novel correlates of immunity and/or protection
The first examples of studies using systems biological tools to understand vaccine induced immune responses came from two independent studies that identified early molecular signatures induced in humans vaccinated with the yellow fever vaccine YF-17D (Gaucher et al., 2008; Querec et al., 2009). YF-17D is a live attenuated vaccine, which was generated after serial passage of a corresponding pathogenic strain (Asibi strain) of the yellow fever virus (Theiler and Smith, 1937), and is one of the most successful vaccines ever developed, as it confers protection in nearly 90% of vaccinees. Over 600 million people have received this vaccine, and a single immunization results in a broad spectrum of immune responses (neutralizing antibodies, cytototoxic T cells, T helper 1 (Th1) and Th2 cells) and neutralizing antibody responses that persist for nearly 4 decades. The goal of our study (Querec et al., 2009) was to use YF-17D as a model to determine the feasibility of applying systems biological approaches to: (a) identifying molecular signatures induced early after vaccination, which could predict the later immunogenicity of the vaccine (i.e. to identify biomarkers of vaccine efficacy); (b) to obtain biological insights about the mechanism of action of YF-17D. Fifteen individuals who had previously not been vaccinated with YF-17D or infected with yellow fever (and were thus immunologically naïve to the vaccine or pathogen) were vaccinated, and blood samples isolated at baseline and at various time points post vaccination, and analyzed with respect to several immunological parameters. There was a striking variation in the magnitude of the antigen-specific CD8+ T cell responses, and the neutralizing antibody titers measured at day 15 or 60, between different individuals (Querec et al., 2009). We then measured cytokine induction in the plasma using a multiplex cytokine assay, and the frequencies and activation status of innate immune cells such as DC and monocyte subsets at days 1,3 or 7 post vaccination, but these measurements did not correlate with the later T cell or antibody responses. Microarray analyses using the Affymetrix Human Genome U133 Plus 2.0 array of total PBMCs revealed a molecular signature comprised of genes involved in innate sensing of viruses and antiviral immunity, in most of the vaccinees. Thus, in addition to enhanced expression of endosomal TLRs, the gene expression of member s of the 2–5 oligoadenylate synthetase family (e.g OAS 1,2,3 and L, which are essential proteins involved in the innate immune response to viral infection), DDX58 (RIG-I) and IFIH1 (MDA-5) were all upregulated (Figure 1). Two key transcription factors that mediate type I interferon responses, IRF7 and STAT1, were also upregulated. Members of the ISGylation pathway, which preserve essential proteins from being degraded during the IFN induced cellular antiviral state, were increased, including ISG15, HERC5, and UBE2L6. Another PRR group where both positive and negative regulation is induced by YF-17D is in the complement cascade. The complement signature of YF-17D included the upregulation of genes for C1q and its feedback inhibitor C1IN and the increased expression of the gene encoding C3a receptor 1 with corresponding increase in the C3a protein in plasma (Figure 1). Thus YF-17D activates multiple pathogen surveillance mechanisms in several cellular compartments: extracellular, cell membrane, cytoplasmic, and vesicular (Figure 1). However, these signatures did not correlate with the magnitude of the antigen-specific CD8+ T cell or antibody responses.
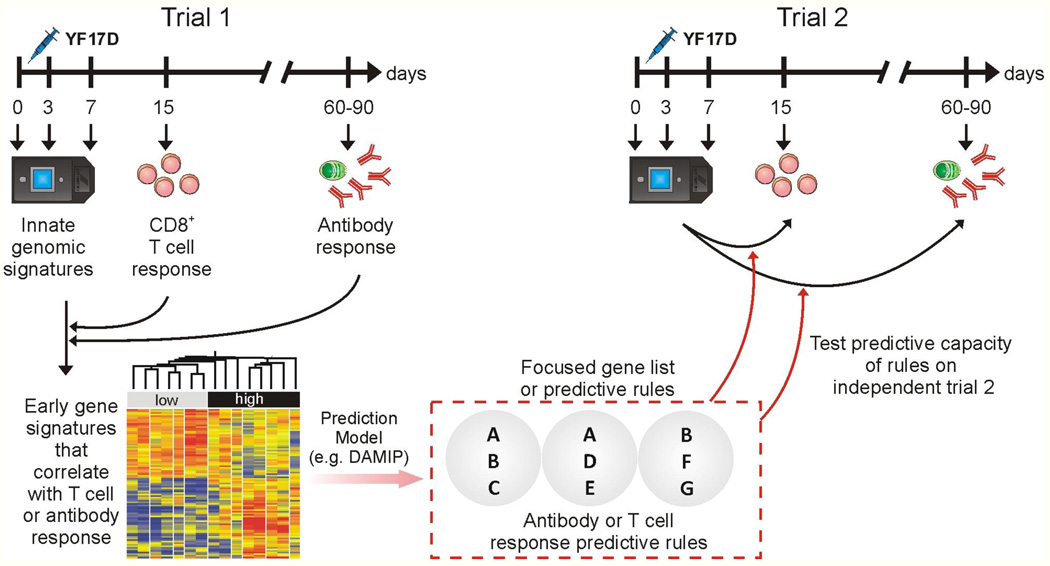
Figure 1. Using Systems Biology to predict the immunogenicity of the YF-17D vaccine.
Schematic representation of the systems biology approach used to predict the T and B cell responses of YF-17D vaccinees (Querec et al., 2009). Healthy humans vaccinated with YF-17D are bled at the indicated time points and the innate and adaptive responses studied. Innate signatures obtained using microarrays are found to correlate with the later adaptive immune responses. The predictive power of such signatures are tested in an independent trial (trial 2).
We then used additional bioinformatics approaches, to identify gene signatures that did correlate with the magnitude of antigen-specific CD8+ T-cell responses and antibody titers, and that were capable of predicting the magnitude of these responses in an independent trial of YF-17D vaccination in humans. We observed signatures for CD8+ T-cell responses from the first trial were predictive with up to 90% accuracy in the second trial and vice versa. Of the genes present in these predictive signatures, EIF2AK4 is known to be a critical player in the integrated stress response (Wek et al., 2006) and regulates protein synthesis in response to changes in amino acid levels by phosphorylating the elongation initiation factor 2 (eIF2α) [Figure 1]. This results in a global shut down of translation of constitutively active proteins, by redirection of their mRNAs from polysomes to discrete cytoplasmic foci known as stress granules (SGs), where they are transiently stored (Kedersha and Anderson, 2007). Consistent with this, YF-17D induced the phosphorylation of eIF2α, and formation of stress granules (Querec et al., 2009). Moreover, several other genes involved in the stress response pathway, like calreriticulin, protein disulfide isomerase, the glucocorticoid receptor and c-Jun, were observed to correlate with the CD8+ T cell response (Figure 1). These observations stimulate the hypothesis that the induction of the integrated stress response in the innate immune system might play a key role in shaping the CD8+ T cell response to YF-17D. Experiments to test the hypothesis are currently underway. In the case of antibody responses, TNFRSF17 (BCMA), a receptor for the B cell growth factor BLyS or BAFF (known to play a key role in B cell differentiation) (Avery et al., 2003), was a key gene in the predictive signatures. Thus, taken together, these studies provide a global description of the innate and adaptive immune responses that are induced after YF17D vaccination and stimulate the generation of testable hypotheses about the biological mechanisms that regulate the magnitude and nature of the immune response to YF-17D (Figure 1).
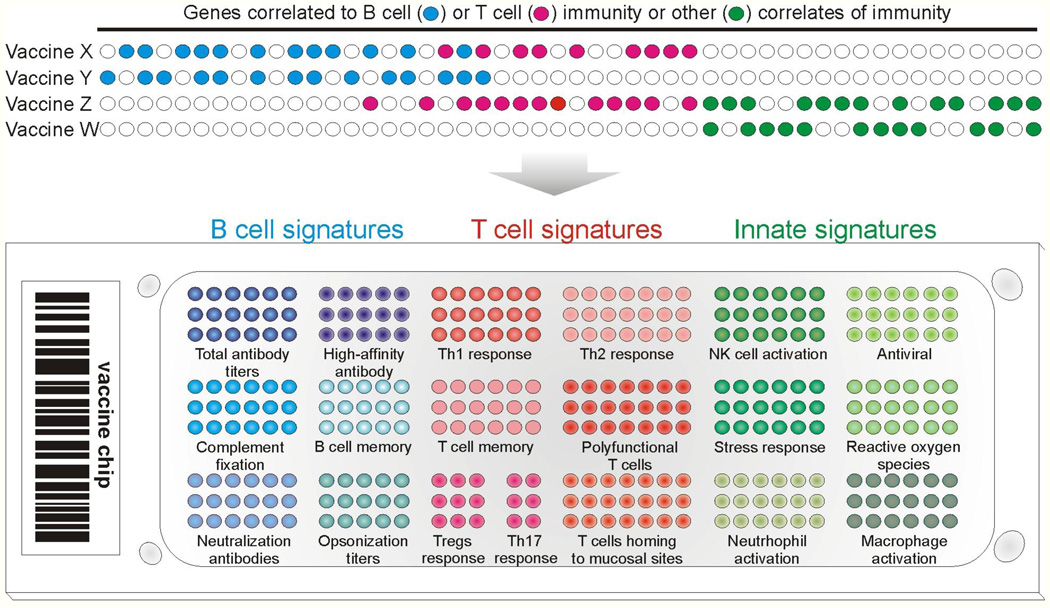
The utility of such an approach in predicting the immunogenicity and protective efficacy of other vaccines need to be determined. The question of whether the signatures that predict the T and B cell responses to YF-17D can also predict such responses to other vaccines remains to be determined. In one scenario it could be envisioned that all vaccines that stimulates antibody responses would induce a common “archetypal” signature, capable of predicting the magnitude of the antibody response to any vaccine. Similarly, there could be an “archetypal” signature that predicts the antigen-specific CD8+ T cell responses to any vaccine. However, B and T cell responses come in different flavors, and different vaccines induce different types of B and T cell responses. It seems unlikely therefore that a common archetypal signature would be capable of predicting all the different types of B or T cell responses induced by different vaccines. A second scenario is that each vaccine could have a very unique signature, which was capable of predicting the particular type of T or B cell responses only to that vaccine. However, many vaccines induce similar types of immune responses (e.g. neutralizing antibodies or polyfunctional CD8+ T cells), so it is reasonable to suggest that vaccines that stimulate a similar mechanism of protective immunity will induce similar molecular signatures. For example, vaccine Y that stimulates long lived plasma cells that produce high affinity antibody may stimulate a particular signature, while vaccine Z that induces polyfunctional CD8+ T cells would stimulate a different signature (Figure 2). Vaccine X that induced both types of responses would stimulate a combined signature (Figure 2). Other vaccines that relied on opsono-phagocytic antibodies for protection may have a different innate signature. Thus, one would have a cluster of signatures that predict various aspects of B cell immunogenicity or T cell immunogenicity. Similarly, there could be a different cluster of signatures that predict protective immunity that is not mediated by T or B cell dependent mechanisms, but by other mechanisms mediated perhaps by NK cells, or DCs, or stress response pathways (Figure 2). In this context, our preliminary data with the influenza vaccines suggest that TNFRSF17, which was a key predictor of the neutralizing antibody responses to YF-17D (Querec et al., 2009), is also a predictor of the hemagglutinin antibody titers to vaccination with the inactivated influenza vaccine, suggesting that there are likely to be common predictors of antibody responses to many vaccines (unpublished data). This probably underlies common biological mechanisms by which different vaccines could stimulate antibody responses. The identification of such predictive signatures will facilitate not only the rapid screening of vaccines, and the development of a vaccine chip, comprising of clusters of a few hundred or fewer genes, each cluster being capable of predicting a facet of immunogenicity (Figure 2B). Such a chip would therefore be used to predict the immunogenicity or virtually any vaccine. Indeed, in the field of cancer genomics, after several years of false starts, “MammaPrint” (http://www.agendia.com/pages/prognosis____prediction/31.php), a prognostic chip for breast cancer, was developed by Agendia, and approved by the Food and Drug Administration in the United States. Like the story behind this breast cancer prognostic chip, the development of the “vaccine chip” will probably require the analysis of hundreds of vaccinees, over several clinical trials. However, we have already seen how host gene expression profiles induced after vaccination correlate with, and predict, vaccine immunogenicity, and also offer mechanistic insights into immune regulation (Querec et al, 2008). This additional layer of knowledge, translated into an array of functional modules on the vaccine chip, gives us extra power that was not utilized in the earlier, brute-force biomarker hunting.
Figure 2. Construction of a Generic Vaccine Chip.
Top: Systems biology approaches allow the identification of predictive gene signatures of immunogenicity for many vaccines. Vaccines with similar correlates of protection may or may not share the same gene markers. The identification of predictive signatures of many vaccines would enable the development of a vaccine chip. Bottom: This chip would consist of perhaps a few hundred genes, subsets of which would predict a particular type of innate or adaptive immune response (e.g. magnitude of effector CD8+ T cell response, frequency of polyfunctional T cells, balance of T helper 1 (Th1), Th2 and Th17 cells, high-affinity antibody titers and so on). This would allow the rapid evaluation of vaccinees for the strength, type, duration and quality of protective immune responses stimulated by the vaccine. Thus, the vaccine chip is a device that could be used to predict immunogenicity and protective capacity of virtually any vaccine in the future.
This is likely to have an impact on several public health related issues in vaccinology. One major issue is that many common vaccines such as the influenza vaccine (Gardner et al., 2006), pneumococcal vaccine (Jackson and Janoff, 2008) and zoster vaccines induce suboptimal immune responses in a substantial proportion of the elderly, or in infants, or in immune compromised populations such as HIV-infected or transplant patients. Therefore, delineation of signatures of immunogenicity would permit such individuals to be identified prospectively. In addition, this strategy will help identify non responders when vaccinating first responders during an emerging outbreak, or when evaluating the efficacy or immunogenicity of untested vaccines (Table 1). Furthermore, the predictive signatures could highlight novel correlates or protective immunity, and enable the formulation of new hypotheses about the mechanisms underlying vaccine induced protective immunity.
Table 1.
Methods to measure antibody correlates of protection (Adapted from Plotkin, 2008)
| Vaccine (Pathogen) | Test | Correlate of Protection |
|---|---|---|
| Diphtheria (C.diphtheriae) | Toxin neutralization | 0.01–0.1 IU/ml |
| Hepatitis A | ELISA | 10 mlU/ml |
| Hepatitis B | ELISA | 10 mlU/ml |
| Hib polysaccharide (Hib) | ELISA | 1 µg/ml |
| Hib conjugate (Hib) | ELISA | 0.15 µg/ml |
| Influenza | HAI | 1/40 dilution |
| Lyme disease | ELISA | 1,100 EIA U/ml |
| Measles | Microneutralization | 120 mlU/ml |
| Pneumococcus (S.pneumoniae) |
ELISA; opsonophagocytosis | 0.2–0.35 µg/ml (for children); 1/8 dilution |
| Polio | Neutralization | 1/4 – 1/8 dilution |
| Rabies | Neutralization | 0.5 IU/ml |
| Rubella | Immunoprecipitation | 10–15 mlU/ml |
| Tetanus | Toxin neutralization | 0.1IU/ml |
| Chickenpox (VZV) | FAMA; gpELISA | ≥ 1/64 dilution; ≥ 5IU/ml |
Historically, correlates of protection have relied on the measurement of the magnitude of the antigen-specific antibody response stimulated by vaccination. Such measurements typically include the concentration of the binding antibody titers (ELISA), or some measure of the activity of the antibody, such neutralization titers or opsonophagocytic titers. When a given threshold of such a measurement is achieved or exceeded, vaccination is assumed to have reached a signature of protective immunization. These tests have become well standardized and relatively straight forward to perform. The name of the pathogen is included in parenthesis, where it’s name is different from the commonly used name for the vaccine.
Abbreviations: C. diphtheria, Corynebacterium diphtheriae; Hib, Haemophilus influenza type B; S.pneumoniae, Streptococcus pneumonia; HAI, hemagglutination inhibition; EIA, enzyme immunoassay; FAMA, fluorescent antibody to membrane antigens; gpELISA, glycoprotein antibody ELISA; VZV, varicella zoster virus.
Systems biology may also be useful in addressing a major challenge in vaccine development: to determine the correlates of protection against a pathogen. The magnitude of the antigen-specific antibody titers is considered to be the primary correlate of protection against most pathogens (Plotkin, 2008) (Table 1). For example, antibodies mediate protection against blood borne viruses such as hepatitis (Jack et al., 1999; Van Damme and Van Herck, 2007) and yellow fever (Lang et al., 1999; Reinhardt et al., 1998; Wheelock and Sibley, 1965), bacteria that secrete toxins that cause diphtheria (Ipsen, 1946) and tetanus (Looney et al., 1956), viruses that infect via mucosal surfaces such as influenza (Dowdle et al., 1973; Mostow et al., 1973) and rotaviruses (Jiang et al., 2008), rabies virus (Nagarajan et al., 2008) which infect neuronal axons, and pneumococcal and meningococcal bacteria, which are leading causes of pneumonia and meningitis (Andreoni et al., 1993; Romero-Steiner et al., 2006). The antigen-specific antibody responses to such vaccines are measured through standardized assays, such as ELISAs (which measure binding antibody titers), hemagglutination inhibition and functional measures of antibody activity such as neutralization and opsonophagocytosis (Table 1). Typically, such assays yield a single value, a threshold, above which antibody responses are considered to be protective.
However, despite the widespread use of such antibody assays to measure the efficacy of current vaccines, in the case of many vaccines humoral immunity may not be the only, or even the best, correlate of protection. Furthermore, protective immunity may not even correlate with the humoral immune response. Varicella virus vaccination efficacy is usually determined by measuring antibody titers using serum neutralization or ELISA. However persistent varicella specific T cells have been shown to be indicators of protection from varicella virus infection and have been suggested as possible additional or alternative correlates of protection in children and the elderly (Arvin, 2008; Levin et al., 2008). Furthermore, antibody titers to influenza vaccination may be unreliable for predicting risk of influenza illness in the elderly population (McElhaney et al., 2006). On the contrary, elderly individuals that have strong influenza-specific T cell responses are less likely to develop flu regardless of post-vaccination antibody titers (McElhaney et al., 2006). Although antibody titers could not distinguish between elderly subjects that did or did not develop flu, those subjects with high IFN-γ:IL-10 ratios following ex vivo stimulation of PBMCs with live influenza preparations, were more likely to be protected from influenza illness (McElhaney et al., 2006). In addition, patients with high frequencies of CMV-specific T cells are less likely to have reactivation of CMV, when they are placed on immunosuppressive drugs to prevent transplant rejection (Bunde et al., 2005; Sester et al., 2001). In fact, many diseases that are a top priority for vaccine development, such as HIV, TB, and malaria, are believed to require strong T cell responses for protection (Hoft, 2008; Pantaleo and Koup, 2004; Reyes-Sandoval et al., 2009). These realizations have led to interest in measuring T cells as correlates of protection.
However, measuring the functional signature of the T cell response as a correlate of protection is more challenging than assessing antibody titers. First, T cell populations are phenotypically and functionally diverse (e.g. CD8+ T cell, CD4+, effector memory, central memory, Th1, Th2, Th17 cells etc). Vaccination can induce the proliferation and differentiation of antigen specific T cells into effector cells that secrete cytokines such as IFNγ, IL-4, IL-17, IL-10, IL-9, or effector memory cells, and central memory cells, all of which play key roles in mediating short and/or long term protective immunity to the pathogen (Harari et al., 2004; Sallusto et al., 1999) (Sallusto, Ahmed, Lanzavecchia – this volume of Immunity). Recent studies have monitored activated T cells in humans, phenotypically by measuring upregulation of CD38 and HLA-DR or peptide-MHC tetramer staining cells (Akondy et al., 2009; Appay et al., 2002; Callan et al., 1998; Morgan et al., 2008). Differentiation into effector and memory phenotypes can be assessed by the expression of markers such as CD45RA, CD62L, CD127, and CCR7 (Akondy et al., 2009; Appay et al., 2002; Callan et al., 1998; Morgan et al., 2008). However, the frequencies of differentiated T cell phenotypes may not be adequate correlates of protection, since these may not necessarily correlate with their functional activity. The functions of T cells can be dependent on the cytokines they secrete (e.g. IFN-γ, IL-2, TNF-α) or production of perforin, as well as other measures of cell proliferation and cell-mediated cytotoxicity. Thus, there are a variety of T cell functional signatures that can be measured as potential correlates of protection, in lieu of the traditional antibody response. Importantly, the assessment of a single parameter of T cell function (e.g. IFNγ secretion) may not be sufficient as a correlate of protection; however, using a functional signature comprised of 2 or more types of measurements may provide more specific and reliable correlates of protection (Harari et al., 2004). Finally it may be necessary to abandon the simple linear functional signature model developed for antibody titers, where a predetermined threshold is used as a correlate. Instead of using a set threshold of a single variable to determine vaccine efficacy, so called “co-correlates of protection” may be more appropriate, where it is the balance among multiple variables that indicates efficacy (Qin et al., 2007). For instance, protection against a pathogen may be achieved when two conditions are satisfied: 1) the frequency of Th1 CD4+ effector memory cells meets a given threshold, and 2) the magnitude of the neutralizing antibody titers reaches a certain threshold. In individuals in whom the thresholds for each of these conditions are not met, it may be the interaction between various co-correlates and not independent levels of each which provides a functional signature of vaccine efficacy. For instance, in the control of viruses or intracellular pathogens the lower the neutralizing antibody titer induced by a vaccine, the higher the cytotoxic T cell response needs to be to enhance the likelihood of protection.
The notion that the innate immune response to vaccination might represent a viable correlate or protection has only recently been considered. Given the pivotal role of the early innate response in regulating the magnitude, quality and duration or the later adaptive immune responses, (Iwasaki and Medzhitov, 2010), specific signatures of innate activation may indicate that the vaccine induced the appropriate quality and sufficient strength of activation to induce protective acquired immunity. As discussed above, it has been shown with yellow fever vaccine 17D that molecular signatures in the blood 3 to 7 days post vaccination, corresponding with vaccine viremia and activation of the innate immune pathways, may be used to predict the peak frequency of activated virus-specific T cells and long term neutralizing antibody titers (Querec et al., 2009).
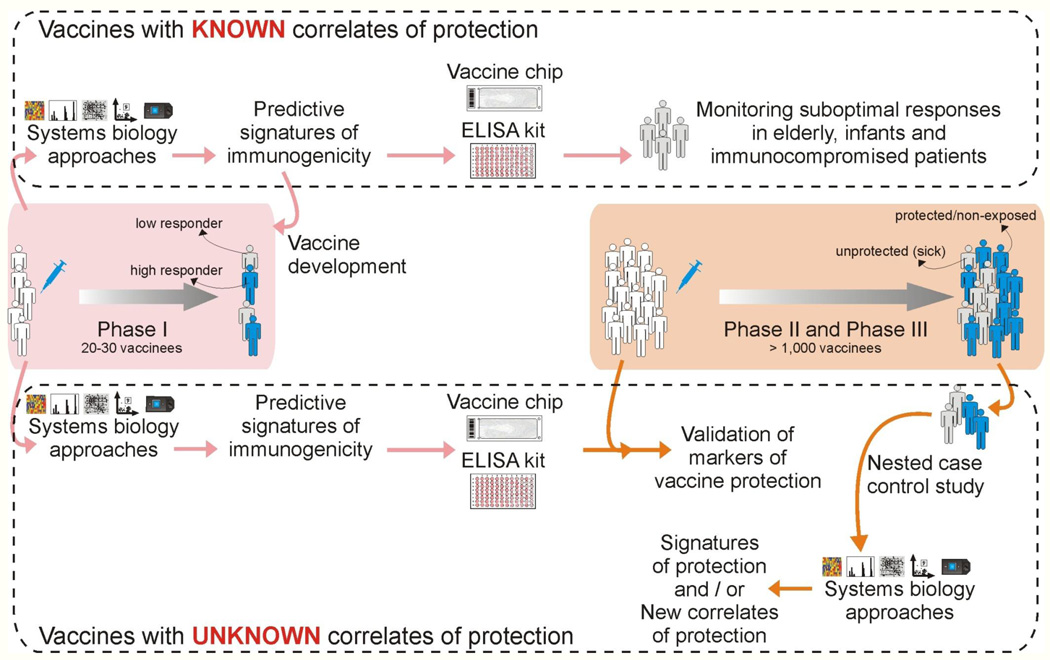
How can systems approaches be integrated into the clinical trial framework, to identify correlates of protective immunity? At the outset, it is important to clarify a frequent source of confusion that arises regarding correlates of immunogenicity versus correlates of protection. The ultimate goal is of course to determine vaccine induced signatures a few hours or days post vaccination that can predict whether a given individual will develop long term protective immunity against the pathogen. The most logical way of addressing this goal is to perform a clinical trial in which vaccinated humans can be challenged with the pathogen, and then to identify signatures that would discriminate between those vaccinees who succumbed to the infection versus those who were protected. With very rare exceptions, as in the case of malaria vaccine trials (Vahey et al., 2010), such an approach is clearly untenable ethically, and thus alternative approaches must be considered. One alternative approach is to use animal challenge models, in which vaccines can be evaluated. Such models, such as the non-human primate model for HIV, or the ferret model for influenza have greatly accelerated vaccine discovery and offered much insight into the mechanisms of protection (Sui et al., 2010). However in some cases, opinions vary regarding the relative merits of a given model, and in translating results obtained from such a model into the clinic (Morgan et al., 2008). Therefore, an alternative or even complementary approach is to identify signatures of immunogenicity to the vaccine, in humans. This approach relies on the axiom that immune protection against a pathogen is mediated by one or more components of the immune response, which can broadly be divided into the adaptive (antigen-specific B and T cells) and the innate responses. Therefore, if there was a priori knowledge of precisely which component(s) of the immune response (e.g. a combination of persistent neutralizing antibody responses and memory CD8+ T cells that migrate to mucosal tissues), then it becomes a relatively straightforward to conduct a phase 0 or1 clinical trial (similar to the yellow fever vaccine trials) in which early predictive signatures of such responses can be identified (Figure 3). Such signatures can then be applied in the clinic to identify vaccinees who will respond sub-optimally to the vaccine. But how do we know what types of immune responses are necessary for protection? In many cases, we can be guided by more than a century of immunological wisdom. For example, few immunologists would deny that the induction of persistent neutralizing antibody responses (Table 1) and cytotoxic T cells are beneficial to fight most viral infections. In such cases, early signatures that various aspects of T or B cell immunogenicity can be assessed in a high throughput manner, using a small number of genes (Vaccine Chip) or an ELISA kit that measured protein expression (Figure 3).
Figure 3. Integrating systems biology approaches into clinical trials.
Top: For vaccines for which correlates or protection are known (Table 1), systems approaches can be used to identify early signatures of protection in a phase 1 trial. The key genes from these signatures can be incorporated into a vaccine chip or ELISA kit, which can then be used to identify non responders or sub-optimal responders, particularly in special populations such as immunocompromised patients, elderly and infants. Bottom: For new and emerging vaccines, for which correlates of protection were unknown, signatures that predict various aspects of immunogenicity (e.g. CD8+ T cell responses or neutralizing antibody responses) can be assessed in phase I trials. Such signatures can then be incorporated into a vaccine chip or ELISA kit that can then be used in phase II and III trials to determine their capacity to predict protection. Alternatively, a retrospective nested case controlled study could be done in a phase II and III trial to identify signatures of protection.
But what happens in situations in which the types of immune responses required for protection are not readily apparent, or where the full range of responses required for optimally effective protection may be unknown? For example in HIV infections, although neutralizing antibodies and cytotoxic T cells are thought to be important (Letvin, 2007), there is much interest in ascertaining whether there are additional mechanisms that might confer protection. Here, it is interesting to consider how systems approaches may be integrated into phase II and III clinical trials, with a view to identifying new correlates of protection. Two approaches to integrating systems approaches into phase II and III trials is shown in Figure 3. Such trials typically involve thousands of participants, and performing high throughput analyses on all would be prohibitively expensive. In one approach, signatures of various aspects of T and B cell immunogenicity can be established in a smaller phase I trial, and these signatures can be incorporated into a relatively cheap and high throughput assay that can be used to predict immunogenicity in phase II and III trials (Figure 3). The assumption here is that some aspect of the T or B cell response will be protective. In a different approach, blood samples could be collected at a few strategic time points (e.g. days 0, 7 post vaccination), straight into RNA lysis buffer and stored for future use. Once the trial was completed, a retrospective nested case control study could be performed using the stored samples, in which, a detailed analyses of innate and adaptive responses could be performed in say 50 vaccinees who acquired the disease and 50 vaccinees who did not. The goal would be to identify signatures induced early on that would discriminate between those who were protected by the vaccine versus those who were not. A caveat with this approach is that one would not know whether those vaccinees who didn’t acquire the disease were actually protected by the vaccine, or simply never encountered the pathogen. However, in many endemic areas of infection (e.g. in a rural area where cholera is endemic and access to clean drinking water is absent), it may be assumed that exposure to infection is high.
A potential benefit of using functional signatures of innate immunity as correlates of protection is that they occur quite early after vaccination compared to the development of memory T cells and antibody responses which can take weeks, months, or years. Being able to determine vaccine efficacy in a short time is useful for many reasons. The current clinical trial format is very lengthy and costly, and usually offers no insights into why a particular vaccine failed. As such clinical trials represent a major rate limiting step in vaccine development. Having a shorter study period increases the probability of retaining all the subjects for the duration of the study, increasing the proportion of subjects that are tracked from vaccination through the final time point. In addition, measuring functional signature of vaccine-induced innate immunity makes high throughput screening of vaccine candidates more feasible. The short duration of time required to measure innate immune activation relative to the endpoints of acquired immunity means: 1) shorter duration to analyze each batch of vaccine candidates, 2) potentially less resources and costs devoted to the early stage analysis of each vaccine candidate, 3) quicker refinement of vaccine formulations and delivery methods, and 4) identifying why a particular vaccine failed (Figure 3).
Apart from lack of inducing sufficient protection, another common reason for vaccines to fail is severe side effects. These side effects are often associated with over activation of certain components of the innate immune system (Gupta et al., 1993; Pulendran et al., 2008). Thus functional signatures of innate immunity may be used to screen adjuvants or as co-correlates of protection along with parameters of acquired immunity for complete vaccines (antigen + adjuvant). Functional signatures may not only help in the design of protective vaccines but may also help to limit the deleterious side effects.
Finally, systems approaches could also yield biological insights about how vaccines work. One area that could benefit from systems approaches is delineation of the mechanisms by which adjuvants work. While the empiric, live attenuated vaccines contain stimuli that activate the innate immune system, and in effect, “act as their own adjuvants,” recombinant vaccines such as the Hepatitis B vaccine need to be administered with exogenous adjuvants. However, in the nearly 250 years since the introduction of vaccination, although a great variety of adjuvants have been proposed, until very recently only alum, described by Glenny in 1926 was globally licensed for human use (De Gregorio et al., 2008; Lindblad, 2004). However alum is a Th2 cell-inducing adjuvant, and does not induce strong Th1 and CTL responses. Thus, there is an urgent need to develop alternative and safe adjuvants that induce different types of immune response that might be optimally effective against different pathogens. Despite its widespread utility, until very recently its mechanism of action has been shrouded in mystery. It has been suggested that alum works by serving as a depot of antigen in the body. It has also been suggested that alum could cause necrosis in the inoculated tissue, which indirectly activates DCs through danger signals in the form of host inflammatory mediators (De Gregorio et al., 2008; Mbow et al., 2010). The details of this mechanism are only recently being revealed. Recently it was demonstrated that alum, signals via the Nalp3 inflammasome (Eisenbarth et al., 2008; Kool et al., 2008; Li et al., 2008). Thus, DCs or macrophages stimulated in vitro with alum plus LPS induced IL-1β and IL-18 in a caspase-1 and Nalp3-dependent manner (Eisenbarth et al., 2008; Kool et al., 2008; Li et al., 2008; McKee et al., 2009). Despite the convincing in vitro studies, the question of whether Nalp3 is required for the adjuvanticity of alum remains controversial, with some studies demonstrating abrogation of antibody responses in Nalp3-deficient (Nlrp3−/−) mice (Eisenbarth et al., 2008; Li et al., 2008), and other studies showing partial or no effect (Kool et al., 2008; McKee et al., 2009). Thus, the mechanisms by which alum induces Th2 responses are poorly understood, and a systems biological approach (e.g. microarray analyses of signatures in response to an alum adjuvanted versus unadjuvanted vaccine), is likely to be useful in providing new insights on the mechanism of action of alum. In this context, Mosca et al performed an elegant study in mice, to assess the molecular and cellular signatures of vaccine adjuvants, including the squalene-based oil-in-water emulsion MF59 (Mosca et al., 2008), which was licensed for human use a decade ago. The molecular mechanism of action and the target cells of alum and MF59 are still unknown. By combining microarray and immunofluorescence analysis, Mosca et al monitored the effects of the adjuvants MF59, CpG, and alum in the mouse muscle. MF59 induced the expression of 891 genes; in contrast CpG and alum regulated 387 and 312 genes, respectively. Interestingly, there was a core set of 168 genes that were modulated by all adjuvants. Although all adjuvants promoted the recruitment of antigen presenting cells, MF59 triggered a more rapid influx of CD11b+ blood cells compared with other adjuvants. Furthermore, MF59 was the most potent inducer of genes encoding cytokines, cytokine receptors, and adhesion molecules involved in leukocyte migration. Intriguingly two genes identified by microarrays, JunB and Ptx3, suggested skeletal muscle as a direct target of MF59. Taken together, the authors’ interpretation of the data suggests that oil-in-water emulsions are efficient human vaccine adjuvants, because they induce an early and strong immunocompetent environment at the injection site by targeting muscle cells. In addition, we have recently applied this approach to identifying a novel mechanism by which adjuvants that induce Th2 responses (e.g. cysteine proteases), program, DCs to stimulate Th2 responses (Tang et al., 2010). This involves the induction of reactive oxygen species (ROS) in DCs, which is critical for the induction of Th2 responses (Tang et al., 2010).
These studies demonstrate the utility of systems approaches in understanding the mechanism of action of adjuvants, and in identifying mechanisms that contribute to their toxicity. In addition, emerging work in innate immunity are revealing the mode of action of many adjuvants. Under the brand name AS04, monophosphoryl lipid A (MPL), an LPS derivative and a TLR4 ligand, is used in combination with alum in Cervarix, GlaxoSmithKline’s recently approved human papillomavirus vaccine (Hennessy et al., 2010). With the growing number of adjuvants at our disposal to mimic natural infections, we need a frame of reference as to how to use them for maximum efficacy. Turning to the functional signatures of innate immunity induced by some of our most successful vaccines is beginning to shed light on this area. For example, YF-17D activates multiple TLRs including TLR 2, 7, 8, and 9, as well as non TLR PRRs such as RIG-I and MDA-5 (Querec et al., 2006; Querec et al., 2009), which results in the activation of plasmacytoid DCs and myeloid DCs. Similar approaches are being applied to understand innate responses to other vectors such as the attenuated pox vectors MVA and NYVAC (Guerra et al., 2007), baculovirus expressed HIV-virus like particles (Buonaguro et al., 2008).
Systems approaches can also shed light on the mechanisms by which vaccines induce a given type of response. As discussed above, one of the key genes in the predictive signature of YF-17D, EIF2AK4 is known to be a critical player in the integrated stress response (Kedersha & Anderson, 2007) and regulate protein synthesis in response to changes in amino acid amounts by phosphorylating the elongation initiation factor 2 (eIF2α) [Figure 1]. Our recent data demonstrate that immunization of mice deficient in EIF2AK4 with YF-17D, results in substantially diminished CD8+ T cell responses (Unpublished data). The precise mechanism of this is under investigation, but this result demonstrates that the integrated stress response plays a key role in regulating adaptive immunity to a viral vaccine.
Finally, it is important to remember that the complex behavior of biological systems cannot be understood by studying parts in isolation (Germain, 2001; Ideker et al., 2001; Kitano, 2002; Weng et al., 1999). Therefore, vaccinologists need to move beyond merely understanding each of the parts of the immune system in isolation, but rather in understanding how the different parts of the immune system interact among themselves. Indeed, a “unified model” of the cellular and molecular mechanisms that vaccine induced protective immunity is likely to result from studying different ‘hierarchies of organization’ with the immune system. In such a hierarchy, the cell can be considered to be the ‘ground level,’ and zooming into the cell to examine innate receptors and signaling networks offers greater conceptual resolution. In contrast, zooming out from the cell, allows more global views of multi-cellular cooperation (e.g. between DC subsets), and the influence of tissue microenvironments (e.g. intestine versus lung) (Pulendran et al., 2010). In addition, the immune system, as with all biological systems, have redundancies, feedback and feed forward regulation, and synergism, which all impact how the instruction of the vaccine are processed (Kitano, 2002). For example, combinatorial triggering of specific combinations of TLRs results in a synergistic production of pro-inflammatory cytokines, via a mechanism dependent on TRIF and MyD88 signaling (Napolitani et al., 2005). Consistent with this, vaccination with nanoparticles containing particular combinations of TLR ligands plus antigens, induced a synergistic enhancement in the magnitude and persistence of antigen-specific memory B cells and long lived plasma cells (Unpublished data).
Low Input, High Throughput, No Output Biology?
Despite the promise of systems approaches in vaccinology, we may do well to heed the advice of Dr. Sydney Brenner (http://sandwalk.blogspot.com/2008/09/in-words-of-sydney-brenner.html; http://www.bio-itworldexpo.com/BioIT_Article.aspx?id=75470&LangType=1033): “The idea that we'll dissect [cellular] complexity by making lots of measurements is bound to fail…Everyone's hoping for a magic computer program - experimental data, pharmacogenomics data, the whole lot - and it will come out with the answer. That's a vague hope. Because I have to tell you, computers are incredibly stupid! It's better to combine human intelligence with artificial stupidity than the other way around …‥ Actually, the orgy of fact extraction in which everybody is currently engaged has, like most consumer economies, accumulated a vast debt. This is a debt of theory, and some of us are soon going to have an exciting time paying it back - with interest, I hope.” The accumulation of a sea of data is but a small stepping stone towards real understanding of biological systems. It is imperative to get beyond colorful heat maps and network maps, to an understanding of the functional significance of the molecular signatures of vaccination. This is a daunting challenge because of several intrinsic problems in this approach. These are discussed below.
Conceptual problems
A major conceptual pitfall lies with the premise that genes that changes in the expression of genes in response to vaccination, may necessarily be functionally relevant for generating the immune response to that vaccine. There are many examples, where genes that are modulated in response are of no consequence to the biological response to that stimulus, because evolution has not had a reason for silencing those genes. Indeed, it is well recognized that gene coexpression only corresponds to causality in very limited cases (Bansal et al., 2007; Schadt et al., 2005). The challenge is to identify true causal relationships among the co-occurring events. One solution is to borrow knowledge from predefined gene modules or pathways. If multiple genes within a module are coordinately regulated by the vaccine, then the likelihood that this module is functionally relevant becomes much higher. Another approach is to combine multiple data types. As Chen et al (Chen et al., 2008) demonstrated, a macrophage-enriched metabolic network, derived by integrating genotyping data and expression data, was causal of obesity traits; while each data type alone could not deliver the predictive power. We should be reminded that the current measurements are still a thin slice of immense biological complexity; microarray data, even with a large sample size, may fail to reach any statistical significance (Dixon et al., 2007). The general question is: how much data, what data, at what resolution, at what scale, are needed to explain the immunological phenotypes? This may be only addressed in each individual case through trials and errors. Finally, the analysis results have to be validated by functional data via proven techniques, say, gene perturbation or deficient mice. As the study design is closely coupled with computational analysis and modeling, systems biology is best done in an environment where biologists and computational scientists interact closely.
A second conceptual problem is the premise that we can deduce mechanistic insights about how the vaccine induced immune responses, by looking at changes in the expression of genes, only in cells isolated from the blood. This is a significant problem, because immune response to local vaccinations will be initiated in the draining lymph nodes. However, with many vaccines, such as live viral or bacterial vectors, there is a transient, systemic replication of the vector and subsequently, a direct activation of blood leukcocytes by the vector. This is likely to produce the profile of gene expression changes observed in the draining lymph nodes, which serves as a surrogate for immunogenicity. Even in the case of non replicating vaccines such as the inactivated influenza vaccine, our result results demonstrate that signatures of immunogenicity can be ascertained in the blood (Unpublished data). An additional problem is that for many vaccines that induce mucosal immunity, gene expression signatures in the blood many not predict the strength, quality and duration of mucosal immunity. Sampling mucosal tissues in human vaccinees is wrought with challenges. Clearly further studies are necessary to ascertain the extent to which immunogenicity of mucosal vaccines can be ascertained from the blood.
Technical problems
One of the key technical issues is that gene expression signatures are prone to artifacts. Since the early studies of cancer expression microarrays, questions have been raised how robust the gene signatures are (Ein-Dor et al., 2006). Recently, emphasis has been placed on pathway and network analyses, as they incorporate prior knowledge into data analysis and are less prone to spurious errors than analyses of individual genes (Chuang et al., 2007; Dinu et al., 2009). This is particularly relevant to immunological studies where signals are often diluted by cell heterogeneity (Haining and Wherry, 2010). In addition, signatures must be validated using additional techniques and independent samples.
Second, when profiling PBMCs, one is looking at signatures from a mixed bag of cells. Therefore, the extent to which the changes in gene expression reflect alterations in the cellular composition of the blood, versus de novo induction of gene expression, remains uncertain. One solution to this problem is to FACS sort subpopulations of cells and then to evaluate expression profiles in individual cell types. However, this approach is rather laborious and expensive. An alternative approach is to devise computational strategies for assessing cell type specific gene expression profiles. Recently, Shen-Orr et al (Shen-Orr et al., 2010) have devised such an approach, using microarray data and relative cell type frequencies. First they validated their approach using predesigned mixtures of cells, and then applied it to whole blood gene expression datasets from stable post-transplant kidney transplant recipients and those experiencing acute rejection.
A third challenge lies in the enormous genetic and environmental heterogeneity in human populations, and the impact that such heterogeneity may have on vaccine induced immunity. Therefore, future studies should strive to conduct such studies in populations that are uniform with respect to age, gender, ethnicity, and immune status. Furthermore, studies that aim to compare vaccine induced immunity between different populations (e.g. frail elderly versus healthy adults) are likely to yield much insights into mechanisms that contribute to impaired immunity in given populations.
Fourth, a major challenge concerns data management and integration of the enormous volume of data generated. The timely sharing of these data is important to the research community. A dedicated database service for vaccine related data, akin to WormBase (Schwarz et al., 2006) and TB database (Reddy et al., 2009), should be created as soon as possible. Public databases for immunology, including InnateDB (Lynn et al., 2008) and Immgen.org (Heng and Painter, 2008), have been well covered by recent reviews (Gardy et al., 2009; Tong and Ren, 2009). In-house databases often become a necessity for high throughput projects. Integration of multiple data types is usually driven by the specific modeling approach, for instance, by naïve Bayesian methods (Huttenhower et al., 2009), by custom algorithms, or combined by biomolecular concepts (Joyce and Palsson, 2006). For example, transcription factor binding data and gene expression are combined under frameworks of transcriptional regulation; metabolites and enzyme expression are combined in metabolic networks. Broader and more definitive immune parameters are desired (Fauci et al., 2008).
Cultural problems
Finally, the successful application of systems approaches to vaccinology requires a close trans-disciplinary collaboration between biologists and computational scientists. It is critical that such individuals engage in active dialogue on a daily basis, to combine rigorous bioinformatics analyses of the data, with biological insights and intuition. Such intimate collaborations could even take place within a single laboratory, where for example, post docs trained in bioinformatics and biology interact closely.
A framework for systems vaccinology
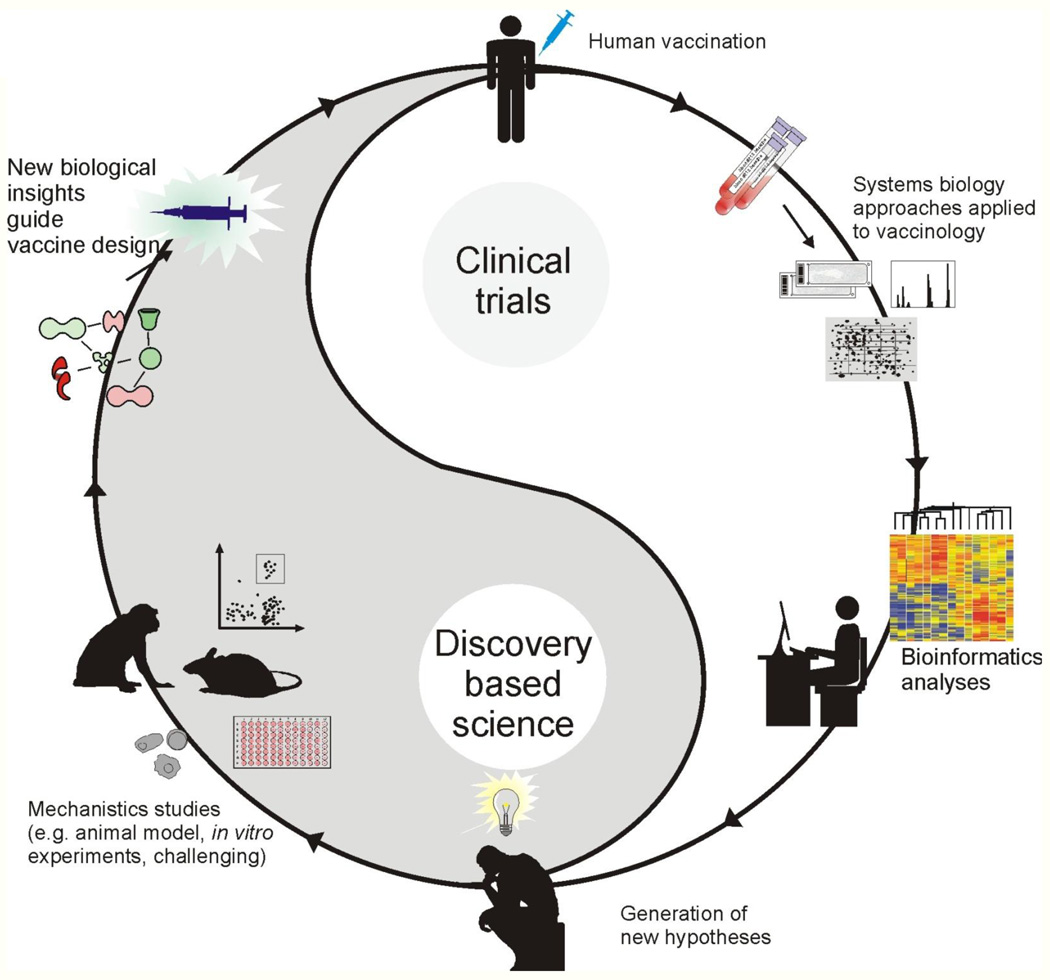
At the World Economic Forum’s annual meeting in Davos this year, Bill Gates pledged $10 billion for vaccines over the next decade, and said that he hoped that the coming ten years would be the “decade of the vaccine.” His words symbolize the unique moment we face today, in our millennial war with pathogens. For the first time, we have begun to understand the mechanisms by which highly successful vaccines mediate protective immunity, and to begin to harness such insights in designing new vaccines against global pandemics. Systems biology promises to offer a new paradigm in vaccinology. Recently, the National Institute of Allergy and Infectious Diseases (NIAID), initiated a new nationwide initiative to establish a consortium of human immune profiling research centers (http://www.eurekalert.org/pub_releases/2010-08/nioa-nle081110.php). The purpose of these centers—which together will receive funding up to $100 million over five years—is to characterize the human immune system under normal conditions and to understand how it changes following infection or vaccination to specific viruses and bacteria. Researchers will use the tools of systems biology to follow the global architecture of the immune response to vaccination or infections in humans, and integrate information about an individual’s genes, proteins and metabolic components, that are perturbed by vaccination or infection. Such studies will be performed in diverse populations with respect to age (including the elderly and children), immune status (including people with autoimmune diseases such as lupus and transplant patients), gender and ethnicity. In addition, the initiative will provide support for centralized infrastructure to collect, characterize, and store the human samples; for bioinformatic capacity to analyze the large and complex data sets that will be generated; and for the discovery and development of new immune response monitoring tools and sample-sparing assays. The results of this initiative are likely to have a major impact on vaccinology, and generate an unprecedented volume of data on immune responses in humans. Yet, we must remember Dr. Brenner’s admonishment and strive to transcend data, and discover knowledge and ultimately understanding. Therefore, the generation of high throughput data represents but a stepping stone towards understanding. An essential aspect of this is to integrate mechanistic studies involving models, both animal and human, (e.g. knockout mice, transgenic mice, siRNA knock down of genes in humans cells in vitro) that can elegantly validate the functions of genes and proteins picked up in the human immune profiling studies (Figure 4). Therefore, data generated in clinical trials can be mined using bioinformatics tools, and used to generate biological hypotheses, which can then be tested using animal models or in vitro systems. The insights gained from experimentation, then guide the design and development of new vaccines (Figure 4). Such a framework seeks to bridge the so called gaps between clinical trials and discovery based science, between human immunology and mouse immunology, between translational and basic science, and offers a seamless continuum of scientific discovery and vaccine invention. That would be emblematic of 21st century vaccinology!
Figure 4. A framework for systems vaccinology.
Systems biology approaches applied to clinical trials can lead to the generation of new hypotheses which can be tested and ultimately lead to developing better vaccines. For example, immune responses to vaccination in clinical trials can be profiled in exquisite depth, using technologies such as microarrays, deep sequencing and proteomics. The high throughput data generated can be mined using bioinformatics tools, and used to create hypotheses about the biological mechanisms underlying vaccine induced immunity. Such hypotheses can then be tested using animal models or in vitro human systems. The insights gained from experimentation, can then guide the design and development of new vaccines. Such a framework seeks to bridge the so called gaps between clinical trials and discovery based science, between human immunology and mouse immunology, between translational and basic science, and offers a seamless continuum of scientific discovery and vaccine invention.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aderem A, Hood L. Immunology in the post-genomic era. Nat Immunol. 2001;2:373–375. doi: 10.1038/87665. [DOI] [PubMed] [Google Scholar]
- Akondy RS, Monson ND, Miller JD, Edupuganti S, Teuwen D, Wu H, Quyyumi F, Garg S, Altman JD, Del Rio C, et al. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol. 2009;183:7919–7930. doi: 10.4049/jimmunol.0803903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Andreoni J, Kayhty H, Densen P. Vaccination and the role of capsular polysaccharide antibody in prevention of recurrent meningococcal disease in late complement component-deficient individuals. J Infect Dis. 1993;168:227–231. doi: 10.1093/infdis/168.1.227. [DOI] [PubMed] [Google Scholar]
- Appay V, Dunbar PR, Callan M, Klenerman P, Gillespie GM, Papagno L, Ogg GS, King A, Lechner F, Spina CA, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8:379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- Arvin AM. Humoral and cellular immunity to varicella-zoster virus: an overview. J Infect Dis. 2008;(197 Suppl 2):S58–S60. doi: 10.1086/522123. [DOI] [PubMed] [Google Scholar]
- Avery DT, Kalled SL, Ellyard JI, Ambrose C, Bixler SA, Thien M, Brink R, Mackay F, Hodgkin PD, Tangye SG. BAFF selectively enhances the survival of plasmablasts generated from human memory B cells. J Clin Invest. 2003;112:286–297. doi: 10.1172/JCI18025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M, Belcastro V, Ambesi-Impiombato A, di Bernardo D. How to infer gene networks from expression profiles. Mol Syst Biol. 2007;3:78. doi: 10.1038/msb4100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunde T, Kirchner A, Hoffmeister B, Habedank D, Hetzer R, Cherepnev G, Proesch S, Reinke P, Volk HD, Lehmkuhl H, et al. Protection from cytomegalovirus after transplantation is correlated with immediate early 1-specific CD8 T cells. J Exp Med. 2005;201:1031–1036. doi: 10.1084/jem.20042384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonaguro L, Monaco A, Arico E, Wang E, Tornesello ML, Lewis GK, Marincola FM, Buonaguro FM. Gene expression profile of peripheral blood mononuclear cells in response to HIV-VLPs stimulation. BMC Bioinformatics. 2008;(9 Suppl 2):S5. doi: 10.1186/1471-2105-9-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan MF, Tan L, Annels N, Ogg GS, Wilson JD, O'Callaghan CA, Steven N, McMichael AJ, Rickinson AB. Direct visualization of antigen-specific CD8+ T cells during the primary immune response to Epstein-Barr virus In vivo. J Exp Med. 1998;187:1395–1402. doi: 10.1084/jem.187.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, Stichweh D, Blankenship D, Li L, Munagala I, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E, Tritto E, Rappuoli R. Alum adjuvanticity: unraveling a century old mystery. Eur J Immunol. 2008;38:2068–2071. doi: 10.1002/eji.200838648. [DOI] [PubMed] [Google Scholar]
- Dinu I, Potter JD, Mueller T, Liu Q, Adewale AJ, Jhangri GS, Einecke G, Famulski KS, Halloran P, Yasui Y. Gene-set analysis and reduction. Brief Bioinform. 2009;10:24–34. doi: 10.1093/bib/bbn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Dowdle WR, Coleman MT, Mostow SR, Kaye HS, Schoenbaum SC. Inactivated influenza vaccines. 2. Laboratory indices of protection. Postgrad Med J. 1973;49:159–163. doi: 10.1136/pgmj.49.569.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci U S A. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth SC, Colegio OR, O'Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci AS, Johnston MI, Dieffenbach CW, Burton DR, Hammer SM, Hoxie JA, Martin M, Overbaugh J, Watkins DI, Mahmoud A, et al. HIV vaccine research: the way forward. Science. 2008;321:530–532. doi: 10.1126/science.1161000. [DOI] [PubMed] [Google Scholar]
- Gardner EM, Gonzalez EW, Nogusa S, Murasko DM. Age-related changes in the immune response to influenza vaccination in a racially diverse, healthy elderly population. Vaccine. 2006;24:1609–1614. doi: 10.1016/j.vaccine.2005.09.058. [DOI] [PubMed] [Google Scholar]
- Gardy JL, Lynn DJ, Brinkman FS, Hancock RE. Enabling a systems biology approach to immunology: focus on innate immunity. Trends Immunol. 2009;30:249–262. doi: 10.1016/j.it.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR, 3rd, Castro E, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijtenbeek TB, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol. 2009;9:465–479. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain RN. The art of the probable: system control in the adaptive immune system. Science. 2001;293:240–245. doi: 10.1126/science.1062946. [DOI] [PubMed] [Google Scholar]
- Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, Kennedy K, Hai T, Bolouri H, Aderem A. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- Guerra S, Najera JL, Gonzalez JM, Lopez-Fernandez LA, Climent N, Gatell JM, Gallart T, Esteban M. Distinct gene expression profiling after infection of immature human monocyte-derived dendritic cells by the attenuated poxvirus vectors MVA and NYVAC. J Virol. 2007;81:8707–8721. doi: 10.1128/JVI.00444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Relyveld EH, Lindblad EB, Bizzini B, Ben-Efraim S, Gupta CK. Adjuvants--a balance between toxicity and adjuvanticity. Vaccine. 1993;11:293–306. doi: 10.1016/0264-410x(93)90190-9. [DOI] [PubMed] [Google Scholar]
- Haining WN, Ebert BL, Subrmanian A, Wherry EJ, Eichbaum Q, Evans JW, Mak R, Rivoli S, Pretz J, Angelosanto J, et al. Identification of an evolutionarily conserved transcriptional signature of CD8 memory differentiation that is shared by T and B cells. J Immunol. 2008;181:1859–1868. doi: 10.4049/jimmunol.181.3.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haining WN, Wherry EJ. Integrating genomic signatures for immunologic discovery. Immunity. 2010;32:152–161. doi: 10.1016/j.immuni.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Harari A, Vallelian F, Pantaleo G. Phenotypic heterogeneity of antigen-specific CD4 T cells under different conditions of antigen persistence and antigen load. Eur J Immunol. 2004;34:3525–3533. doi: 10.1002/eji.200425324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- Hennessy EJ, Parker AE, O'Neill LA. Targeting Toll-like receptors: emerging therapeutics? Nat Rev Drug Discov. 2010;9:293–307. doi: 10.1038/nrd3203. [DOI] [PubMed] [Google Scholar]
- Hoft DF. Tuberculosis vaccine development: goals, immunological design, and evaluation. Lancet. 2008;372:164–175. doi: 10.1016/S0140-6736(08)61036-3. [DOI] [PubMed] [Google Scholar]
- Huttenhower C, Haley EM, Hibbs MA, Dumeaux V, Barrett DR, Coller HA, Troyanskaya OG. Exploring the human genome with functional maps. Genome Res. 2009;19:1093–1106. doi: 10.1101/gr.082214.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyduke DR, Palsson BO. Towards genome-scale signalling-network reconstructions. Nat Rev Genet. 2010;11:297–307. doi: 10.1038/nrg2750. [DOI] [PubMed] [Google Scholar]
- Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- Ipsen J. Circulating antitoxin at the onset of diphtheria in 425 patients. J Immunol. 1946;54:325–347. [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack AD, Hall AJ, Maine N, Mendy M, Whittle HC. What level of hepatitis B antibody is protective? J Infect Dis. 1999;179:489–492. doi: 10.1086/314578. [DOI] [PubMed] [Google Scholar]
- Jackson LA, Janoff EN. Pneumococcal vaccination of elderly adults: new paradigms for protection. Clin Infect Dis. 2008;47:1328–1338. doi: 10.1086/592691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Gentsch JR, Glass RI. Inactivated rotavirus vaccines: a priority for accelerated vaccine development. Vaccine. 2008;26:6754–6758. doi: 10.1016/j.vaccine.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Joyce AR, Palsson BO. The model organism as a system: integrating 'omics' data sets. Nat Rev Mol Cell Biol. 2006;7:198–210. doi: 10.1038/nrm1857. [DOI] [PubMed] [Google Scholar]
- Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- Kitano H. Computational systems biology. Nature. 2002;420:206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- Kool M, Petrilli V, De Smedt T, Rolaz A, Hammad H, van Nimwegen M, Bergen IM, Castillo R, Lambrecht BN, Tschopp J. Cutting edge: alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J Immunol. 2008;181:3755–3759. doi: 10.4049/jimmunol.181.6.3755. [DOI] [PubMed] [Google Scholar]
- Lang J, Zuckerman J, Clarke P, Barrett P, Kirkpatrick C, Blondeau C. Comparison of the immunogenicity and safety of two 17D yellow fever vaccines. Am J Trop Med Hyg. 1999;60:1045–1050. doi: 10.4269/ajtmh.1999.60.1045. [DOI] [PubMed] [Google Scholar]
- Lee E, Chuang HY, Kim JW, Ideker T, Lee D. Inferring pathway activity toward precise disease classification. PLoS Comput Biol. 2008;4:e1000217. doi: 10.1371/journal.pcbi.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letvin NL. Correlates of immune protection and the development of a human immunodeficiency virus vaccine. Immunity. 2007;27:366–369. doi: 10.1016/j.immuni.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Levin MJ, Oxman MN, Zhang JH, Johnson GR, Stanley H, Hayward AR, Caulfield MJ, Irwin MR, Smith JG, Clair J, et al. Varicella-zoster virus-specific immune responses in elderly recipients of a herpes zoster vaccine. J Infect Dis. 2008;197:825–835. doi: 10.1086/528696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Willingham SB, Ting JP, Re F. Cutting edge: inflammasome activation by alum and alum's adjuvant effect are mediated by NLRP3. J Immunol. 2008;181:17–21. doi: 10.4049/jimmunol.181.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad EB. Aluminium compounds for use in vaccines. Immunol Cell Biol. 2004;82:497–505. doi: 10.1111/j.0818-9641.2004.01286.x. [DOI] [PubMed] [Google Scholar]
- Looney JM, Edsall G, Ipsen J, Jr, Chasen WH. Persistence of antitoxin levels after tetanus-toxoid inoculation in adults, and effect of a booster dose after various intervals. N Engl J Med. 1956;254:6–12. doi: 10.1056/NEJM195601052540102. [DOI] [PubMed] [Google Scholar]
- Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan TH, Shah N, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol Syst Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbow ML, De Gregorio E, Valiante NM, Rappuoli R. New adjuvants for human vaccines. Curr Opin Immunol. 2010;22:411–416. doi: 10.1016/j.coi.2010.04.004. [DOI] [PubMed] [Google Scholar]
- McElhaney JE, Xie D, Hager WD, Barry MB, Wang Y, Kleppinger A, Ewen C, Kane KP, Bleackley RC. T cell responses are better correlates of vaccine protection in the elderly. J Immunol. 2006;176:6333–6339. doi: 10.4049/jimmunol.176.10.6333. [DOI] [PubMed] [Google Scholar]
- McKee AS, Munks MW, MacLeod MK, Fleenor CJ, Van Rooijen N, Kappler JW, Marrack P. Alum induces innate immune responses through macrophage and mast cell sensors, but these sensors are not required for alum to act as an adjuvant for specific immunity. J Immunol. 2009;183:4403–4414. doi: 10.4049/jimmunol.0900164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C, Marthas M, Miller C, Duerr A, Cheng-Mayer C, Desrosiers R, Flores J, Haigwood N, Hu SL, Johnson RP, et al. The use of nonhuman primate models in HIV vaccine development. PLoS Med. 2008;5:e173. doi: 10.1371/journal.pmed.0050173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca F, Tritto E, Muzzi A, Monaci E, Bagnoli F, Iavarone C, O'Hagan D, Rappuoli R, De Gregorio E. Molecular and cellular signatures of human vaccine adjuvants. Proc Natl Acad Sci U S A. 2008;105:10501–10506. doi: 10.1073/pnas.0804699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostow SR, Schoenbaum SC, Dowdle WR, Coleman MT, Kaye HS. Inactivated vaccines. 1. Volunteer studies with very high doses of influenza vaccine purified by zonal ultracentrifugation. Postgrad Med J. 1973;49:152–158. doi: 10.1136/pgmj.49.569.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan T, Rupprecht CE, Dessain SK, Rangarajan PN, Thiagarajan D, Srinivasan VA. Human monoclonal antibody and vaccine approaches to prevent human rabies. Curr Top Microbiol Immunol. 2008;317:67–101. doi: 10.1007/978-3-540-72146-8_3. [DOI] [PubMed] [Google Scholar]
- Napolitani G, Rinaldi A, Bertoni F, Sallusto F, Lanzavecchia A. Selected Tolllike receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat Immunol. 2005;6:769–776. doi: 10.1038/ni1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaegui D, Baranzini SE, Armananzas R, Calvo B, Munoz-Culla M, Khankhanian P, Inza I, Lozano JA, Castillo-Trivino T, Asensio A, et al. Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS One. 2009;4:e6309. doi: 10.1371/journal.pone.0006309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo G, Koup RA. Correlates of immune protection in HIV-1 infection: what we know, what we don't know, what we should know. Nat Med. 2004;10:806–810. doi: 10.1038/nm0804-806. [DOI] [PubMed] [Google Scholar]
- Pascual V, Chaussabel D, Banchereau J. A genomic approach to human autoimmune diseases. Annu Rev Immunol. 2010;28:535–571. doi: 10.1146/annurev-immunol-030409-101221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R, Cragun J, Cottrill H, Kelley MJ, Petersen R, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- Pulendran B, Ahmed R. Translating innate immunity into immunological memory: implications for vaccine development. Cell. 2006;124:849–863. doi: 10.1016/j.cell.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Pulendran B, Miller J, Querec TD, Akondy R, Moseley N, Laur O, Glidewell J, Monson N, Zhu T, Zhu H, et al. Case of yellow fever vaccine--associated viscerotropic disease with prolonged viremia, robust adaptive immune responses, and polymorphisms in CCR5 and RANTES genes. J Infect Dis. 2008;198:500–507. doi: 10.1086/590187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Tang H, Manicassamy S. Programming dendritic cells to induce T(H)2 and tolerogenic responses. Nat Immunol. 2010;11:647–655. doi: 10.1038/ni.1894. [DOI] [PubMed] [Google Scholar]
- Qin L, Gilbert PB, Corey L, McElrath MJ, Self SG. A framework for assessing immunological correlates of protection in vaccine trials. J Infect Dis. 2007;196:1304–1312. doi: 10.1086/522428. [DOI] [PubMed] [Google Scholar]
- Querec T, Bennouna S, Alkan S, Laouar Y, Gorden K, Flavell R, Akira S, Ahmed R, Pulendran B. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J Exp Med. 2006;203:413–424. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TB, Riley R, Wymore F, Montgomery P, DeCaprio D, Engels R, Gellesch M, Hubble J, Jen D, Jin H, et al. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 2009;37:D499–D508. doi: 10.1093/nar/gkn652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt B, Jaspert R, Niedrig M, Kostner C, L'Age-Stehr J. Development of viremia and humoral and cellular parameters of immune activation after vaccination with yellow fever virus strain 17D: a model of human flavivirus infection. J Med Virol. 1998;56:159–167. doi: 10.1002/(sici)1096-9071(199810)56:2<159::aid-jmv10>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Reyes-Sandoval A, Pearson FE, Todryk S, Ewer K. Potency assays for novel T-cell-inducing vaccines against malaria. Curr Opin Mol Ther. 2009;11:72–80. [PubMed] [Google Scholar]
- Romero-Steiner S, Frasch CE, Carlone G, Fleck RA, Goldblatt D, Nahm MH. Use of opsonophagocytosis for serological evaluation of pneumococcal vaccines. Clin Vaccine Immunol. 2006;13:165–169. doi: 10.1128/CVI.13.2.165-169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz EM, Antoshechkin I, Bastiani C, Bieri T, Blasiar D, Canaran P, Chan J, Chen N, Chen WJ, Davis P, et al. WormBase: better software, richer content. Nucleic Acids Res. 2006;34:D475–D478. doi: 10.1093/nar/gkj061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sester M, Sester U, Gartner B, Heine G, Girndt M, Mueller-Lantzsch N, Meyerhans A, Kohler H. Levels of virus-specific CD4 T cells correlate with cytomegalovirus control and predict virus-induced disease after renal transplantation. Transplantation. 2001;71:1287–1294. doi: 10.1097/00007890-200105150-00018. [DOI] [PubMed] [Google Scholar]
- Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, Hastie T, Sarwal MM, Davis MM, Butte AJ. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7:287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM. Dendritic cells in vivo: a key target for a new vaccine science. Immunity. 2008;29:319–324. doi: 10.1016/j.immuni.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Sui Y, Zhu Q, Gagnon S, Dzutsev A, Terabe M, Vaccari M, Venzon D, Klinman D, Strober W, Kelsall B, et al. Innate and adaptive immune correlates of vaccine and adjuvant-induced control of mucosal transmission of SIV in macaques. Proc Natl Acad Sci U S A. 2010;107:9843–9848. doi: 10.1073/pnas.0911932107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Cao W, Kasturi SP, Ravindran R, Nakaya HI, Kundu K, Murthy N, Kepler TB, Malissen B, Pulendran B. The T helper type 2 response to cysteine proteases requires dendritic cell-basophil cooperation via ROS-mediated signaling. Nat Immunol. 2010;11:608–617. doi: 10.1038/ni.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theiler M, Smith HH. The Use of Yellow Fever Virus Modified by in Vitro Cultivation for Human Immunization. J Exp Med. 1937;65:787–800. doi: 10.1084/jem.65.6.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JP, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science. 2010;327:286–290. doi: 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong JC, Ren EC. Immunoinformatics: Current trends and future directions. Drug Discov Today. 2009;14:684–689. doi: 10.1016/j.drudis.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahey MT, Wang Z, Kester KE, Cummings J, Heppner DG, Jr, Nau ME, Ofori-Anyinam O, Cohen J, Coche T, Ballou WR, et al. Expression of genes associated with immunoproteasome processing of major histocompatibility complex peptides is indicative of protection with adjuvanted RTS,S malaria vaccine. J Infect Dis. 2010;201:580–589. doi: 10.1086/650310. [DOI] [PubMed] [Google Scholar]
- Van Damme P, Van Herck K. A review of the long-term protection after hepatitis A and B vaccination. Travel Med Infect Dis. 2007;5:79–84. doi: 10.1016/j.tmaid.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Weng G, Bhalla US, Iyengar R. Complexity in biological signaling systems. Science. 1999;284:92–96. doi: 10.1126/science.284.5411.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelock EF, Sibley WA. Circulating Virus, Interferon and Antibody after Vaccination with the 17-D Strain of Yellow-Fever Virus. N Engl J Med. 1965;273:194–198. doi: 10.1056/NEJM196507222730404. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak DE, Aderem A. Systems biology of innate immunity. Immunol Rev. 2009;227:264–282. doi: 10.1111/j.1600-065X.2008.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]