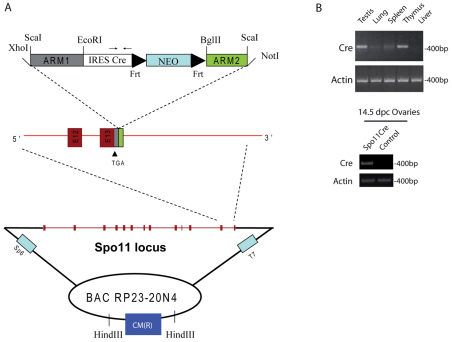
Fig. 1.
Generation and evaluation of Spo11-IRES-Cre mice. (A) BAC targeting of the murine Spo11 locus after the stop codon of the gene. The DNA fragment containing the homology regions ARM1 and ARM2 for the DNA recombination and the IRES-Cre-Neomycin (Neo) sequences was obtained by ScaI enzymatic digestion from the modified PL459 plasmid. The restriction enzymes used to insert the homology regions into the original PL459 plasmid are indicated. Spo11 exons are shown as red boxes, and exons 12 and 13 are magnified. Frt sites flanking the Neo cassette are shown. In the IRES-Cre sequence the location of primers used for Cre analyses is shown. CM(R) represents the chloramphenicol resistance gene used as a probe to identify founder mice. (B) Representative RT-PCR analysis of testis, lung, liver, spleen and thymus mRNA isolated from a 2-month-old mouse of the D5 founder line (top panel) and ovaries mRNA from 14.5 d.p.c. embryo (bottom panel), showing tissue-specific Cre mRNA expression. Actin PCR was used as a loading control.