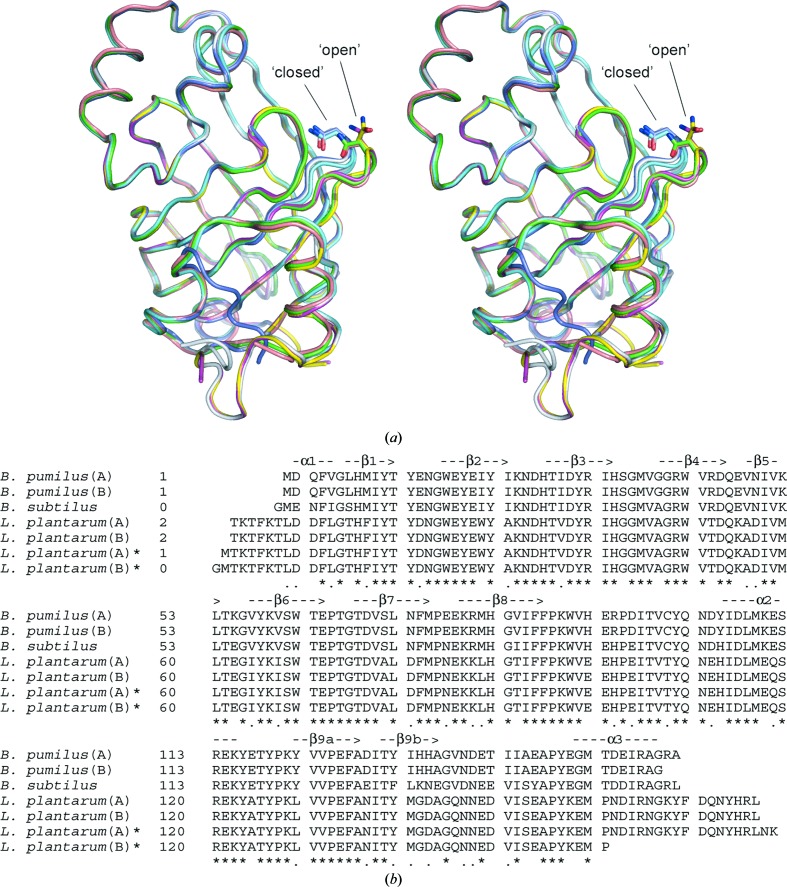
Figure 3.
(a) Structural superposition of PAD subunits from B. pumilus (PDB code 3nad; this study; chain A in green, chain B in cyan), L. plantarum [PDB codes 2w2a (chain A in magenta, chain B in yellow) and 2gc9 (chain A in light blue, chain B in dark blue)] and B. subtilus (PDB code 2p8g; pink). The differences in structure of the β1–β2 loop, which contains a conserved Asn residue and adopts either a ‘closed’ or ‘open’ conformation, are shown. (b) Structure-based sequence alignment of PAD subunits based on the structure superposition shown in (a). Identical residues (*) and residues with similar properties (.) are indicated below the alignment. Secondary-structure elements based on the B. pumilus crystal structure are shown above the alignment. The structural superposition and structure-based alignment were generated using Swiss-PdbViewer (Guex & Peitsch, 1997 ▶).