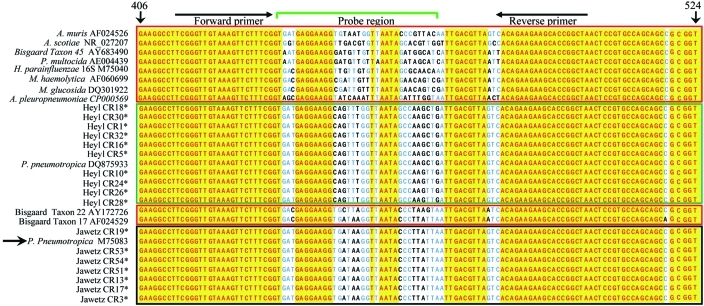
Figure 5.
Alignment of the PCR target region of 16S rRNA from Heyl, Jawetz, and Pasteurellaceae–other. Sequences were aligned by using Vector NTI software; isolates with identical 16S rRNA and rpoB genes have been omitted from the alignment. *, P. pneumotropica isolates sequenced during the current study; green box, Heyl isolates (those sequenced during the current study and DQ875933); black box, Jawetz isolates (those sequenced during the current study and M75083); red boxes, Pasteurellaceae–other sequences; yellow, conserved residues; blue and black letters, mismatches. The sequences have been aligned to nucleotides 406 through 524 of reference strain M75083. The reference strain and nucleotide positions of the reference strain are indicated by arrows. The positions of primers and probes used are indicated on top. The sequence data suggest that although 16S rRNA is highly conserved between various species and biotypes of Pasteurellaceae, small regions unique to P. Heyl and Jawetz biotypes are present and offer an opportunity to design a P. pneumotropica-specific PCR-based diagnostic assay.