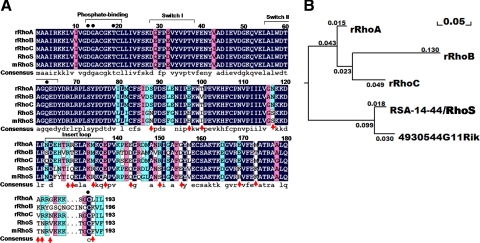
Figure 3.
The structural features distinguish RhoS from canonical Rho GTPases. (A) Alignment of the amino acid sequence of RhoS and three classical Rho GTPases (RhoA, RhoB, and RhoC). The alignment was performed by DNAMAN (Lynnon, Quebec, Canada). Homology levels are highlighted in different colors. Black: 100%; Pink: 75%; Blue: 50%. Arrows: residues conserved between RhoS and 4930544G11Rik (mRhoS) but significantly distinct from RhoA/B/C; Points: conserved resides for regulating the activity of Rho type GTPases. (B) Phylogenetic tree analysis of three Rho isoforms, RhoS, and 4930544G11Rik. Data generated from alignment (Figure 3A) were loaded to reconstruct the rooted neighbor-joining phylogenetic tree by the maximum likelihood method. Numbers indicate branch length. r, rat.