A C-terminal region in Zds2p (ZH4) is required for regulation of Swe1p-dependent polarized cell growth and this region is necessary and sufficient for interaction with protein phosphatase 2A regulatory subunit, Cdc55p. Our results indicate that the Zds proteins regulate the Swe1p-dependent G2/M checkpoint in a CDC55-dependent manner.
Abstract
Deletion of the paralogs ZDS1 and ZDS2 in the budding yeast Saccharomyces cerevisiae causes a mis-regulation of polarized cell growth. Here we show a function for these genes as regulators of the Swe1p (Wee1p) kinase–dependent G2/M checkpoint. We identified a conserved domain in the C-terminus of Zds2p consisting of amino acids 813–912 (hereafter referred to as ZH4 for Zds homology 4) that is required for regulation of Swe1p-dependent polarized bud growth. ZH4 is shown by protein affinity assays to be necessary and sufficient for interaction with Cdc55p, a regulatory subunit of protein phosphatase 2A (PP2A). We hypothesized that the Zds proteins are in a pathway that negatively regulates the Swe1p-dependent G2/M checkpoint via Cdc55p. Supporting this model, deletion of CDC55 rescues the aberrant bud morphology of a zds1Δzds2Δ strain. We also show that expression of ZDS1 or ZDS2 from a strong galactose-inducible promoter can induce mitosis even when the Swe1p-dependent G2/M checkpoint is activated by mis-organization of the actin cytoskeleton. This negative regulation requires the CDC55 gene. Together these data indicate that the Cdc55p/Zds2p module has a function in the regulation of the Swe1p-dependent G2/M checkpoint.
INTRODUCTION
Proper cell division requires a carefully orchestrated progression of cell growth, DNA replication, mitosis, and cytokinesis. Among the cell cycle mechanisms that regulate the progression of these events are checkpoints that arrest or retard the cell cycle when activated in response to cellular damage or perturbation. The entry into mitosis, for example, is regulated by a checkpoint at G2/M that is a key DNA damage and cell size surveillance step. This checkpoint is mis-regulated in many tumors, allowing for tumor cell growth even in the presence of damaged DNA content such as DNA breaks, chromosomal fusions, or aneuploidy (reviewed in Hook et al., 2007; Lobrich and Jeggo, 2007; Bucher and Britten, 2008). In vertebrates and the fission yeast Schizosaccharomyces pombe, the G2/M checkpoint is thought to be triggered by DNA damage (Dasso and Newport, 1990; Enoch and Nurse, 1990) or insufficient cell size (Nurse, 1975). In Saccharomyces cerevisiae, however, the G2/M checkpoint does not monitor DNA damage (Amon et al., 1992; Sorger and Murray, 1992), although recent work has revealed that the DNA replication checkpoint controlled by Rad53p cross-talks with the G2/M checkpoint (Enserink et al., 2006). The S. cerevisiae G2/M checkpoint is active in cells that fail to form a bud, leading to the idea that it is a bud morphogenesis checkpoint (Lew and Reed, 1995). This hypothesis was supported by the fact that extended activation of this checkpoint led to an elongated bud phenotype (Booher et al., 1993) and that loss of checkpoint proteins rescues certain bud morphology mutants (Wang and Burke, 1997; McMillan et al., 1999b). It was shown subsequently, in an elegant set of experiments, that the checkpoint is largely indifferent to bud morphogenesis but will respond to disruption of the actin cytoskeleton (McNulty and Lew, 2005). The link between actin and the G2/M checkpoint is still largely not understood but provides an interesting avenue to further explore how the entry into mitosis is regulated.
The core molecular mechanism for the regulation of the G2/M transition in S. cerevisiae involves the inhibitory phosphorylation of a cyclin dependent kinase (Cdc28p) by Swe1p kinase (see Figure 1 in Results; Booher et al., 1993). This phosphorylation holds the mitosis-inducing Cdc28p/Clb2p (cyclin B) complex in a premitotic state until the inhibitory phosphate is removed by the phosphatase Mih1p. Regulation of the synthesis and degradation of Swe1p kinase (Sia et al., 1996, 1998) as well as the activity of the Mih1p phosphatase (Russell et al., 1989; Dunphy and Kumagai, 1991; Gautier et al., 1991; Kumagai and Dunphy, 1992) are key to cell cycle progression through G2/M. Swe1p is synthesized in late G1 (Sia et al., 1996). As cells approach mitosis Swe1p is hyperphosphorylated and degraded, allowing for dephosphorylation (and thus activation) of Cdc28p/Clb2p (Gould and Nurse, 1989; Lundgren et al., 1991). The efficient phosphorylation and degradation of Swe1p requires the action of Nim1-related kinase Hsl1p (Ma et al., 1996; Tanaka and Nojima, 1996) and the arginine methyltransferase Hsl7p (McMillan et al., 1999a; Miranda et al., 2006; Sayegh and Clarke, 2008). Overexpression of Hsl7p or Hsl1p forces cells to enter mitosis even when the checkpoint is activated by actin depolarization (McMillan et al., 1999a). In contrast to Swe1p, Mih1p regulation is not as well defined. In S. pombe and vertebrate cells, Mih1p (Cdc25p in these organisms) becomes hyperphosphorylated and more active toward Cdc28p (Cdc2p in these organisms) as cells approach mitosis (Moreno et al., 1990; Kumagai and Dunphy, 1992). However, in S. cerevisiae, Mih1p exists in a hyperphosphorylated form throughout the cell cycle until cells approach mitosis when it is dephosphorylated by protein phosphatase 2A (PP2A; Pal et al., 2008). Which phosphoform of Mih1p in S. cerevisiae is more active toward Cdc28p/Clb2p has yet to be defined.
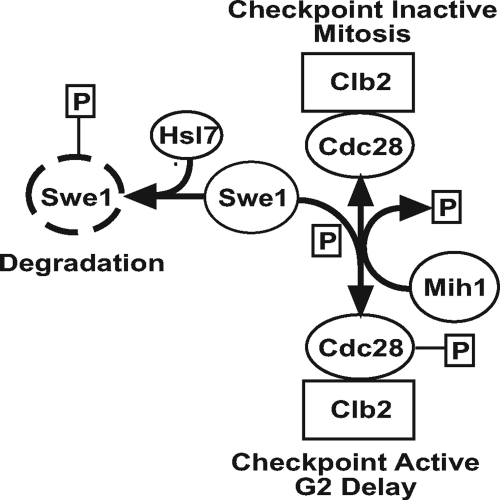
Figure 1.
Core mechanism of the Swe1p-dependent G2/M checkpoint in S. cerevisiae. Mitotic progression requires degradation of Swe1p kinase. Failure to degrade Swe1p kinase results in an inhibitory phosphorylation of the cyclin-dependent kinase Cdc28p and activation of the Swe1p-dependent G2/M checkpoint, delaying mitosis. See Introduction for a full description of the checkpoint mechanism.
PP2A is a heterotrimeric dual specificity serine/threonine phosphatase (structure reviewed in Janssens et al., 2008; Eichhorn et al., 2009) consisting of a core enzyme with a catalytic C subunit and a structural A subunit. Substrate specificity is determined by a regulatory B subunit of which S. cerevisiae has two: the B subunit Cdc55p (Shu et al., 1997) and the B′ subunit Rts1p (Wang and Burke, 1997). PP2A was identified as a major tumor suppressor when okadaic acid, an inhibitor of PP2A, was shown to cause tumors in mice (Bialojan and Takai, 1988; Suganuma et al., 1988). Mutation of, and mis-regulation by, the B subunits account for many of the known disruptions of PP2A-mediated tumor suppression (Eichhorn et al., 2009). In S. cerevisiae, deletion of CDC55 leads to a constitutively hyperphosphorylated form of the phosphatase, Mih1p, which removes the inhibitory phosphate on Cdc28p/Clb2p (Pal et al., 2008). Additionally, CDC55 deletion causes a cold-sensitive mutant bud morphogenesis phenotype (Healy et al., 1991), which is checkpoint dependent (Wang and Burke, 1997), indicating that CDC55 is involved in regulation of the Swe1p-dependent G2/M checkpoint.
S. cerevisiae provides a valuable opportunity to discover novel mechanisms of Swe1p (Wee1p)-dependent G2/M checkpoint regulation. Two paralogous genes, ZDS1 and ZDS2, are of particular interest in this regard. Deletion of these genes results in an SWE1-dependent elongated bud morphology (Wang and Burke, 1997; McMillan et al., 1999b). The ZDS (Zillion Different Screens) genes have been linked by a multitude of genetic screens to various cellular processes including; telomeric silencing (Roy and Runge, 1999; Hsu et al., 2004), transcriptional silencing (Roy and Runge, 1999, 2000; Pierstorff and Kane, 2004), mitotic exit (Heo et al., 1999; Queralt and Uhlmann, 2008), mRNA export (Griffioen et al., 2001; Estruch et al., 2005), regulation of the Rho family GTPase Cdc42 (Bi and Pringle, 1996), calcium sensitivity (Yokoyama et al., 2006; Ohnuki et al., 2007), cell cycle regulation (Yu et al., 1996), cell wall integrity (Griffioen et al., 2001; Sekiya-Kawasaki et al., 2002; Zanelli and Valentini, 2005; Yakura et al., 2006), and others (Ma et al., 1996; Schwer et al., 1998; Bandhakavi et al., 2003). It is not known whether deletion of the ZDS genes disrupts one or more of these functions, triggering the Swe1p-dependent G2/M checkpoint or whether the Zds proteins can directly regulate the Swe1p-dependent G2/M checkpoint.
As the first step toward deciphering how the Zds proteins interface with the Swe1p-dependent G2/M checkpoint, we structure–function mapped Zds2p. We identified a conserved C-terminal domain of Zds2p as necessary for the regulation of bud growth. We also show that this domain is necessary for interaction with Cdc55p, a PP2A regulatory subunit. Our data indicate that CDC55 is necessary for elongated bud growth in cells lacking ZDS1 and ZDS2. Furthermore, we show that overexpression of Zds1p and Zds2p down-regulates the Swe1p-dependent G2/M checkpoint and this regulation requires CDC55. These data lead us to propose a model in which the Zds proteins regulate the Swe1p checkpoint in a Cdc55p-dependent manner.
MATERIALS AND METHODS
Strains and Microbial Techniques
Yeast strains and plasmids used in this study are listed in Tables 1 and 2. Yeast were grown at 25°C unless indicated otherwise. S. cerevisiae of the S288C background were grown in rich medium (YPD) or in synthetic complete medium (SC) lacking a specific amino acid or uracil (Sherman et al., 1986). Strains of the BF264–15DU background (used in GAL induction experiments) were grown as described below. All yeast media contained 2% glucose as a carbon source unless otherwise specified. Yeast transformations were performed as in Kozminski et al. (2006).
Table 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| DLY690 | MATa cdc24-1 swe1::LEU2 bar1 ade1 his2 leu2-3,112 trp1 ura3Δns | McMillan et al. (1999b)a,b |
| DLY657 | MATa cdc24-1 bar1 ade1 his2 leu2-3,112 trp1 ura3Δns | McMillan et al. (1999b)a,b |
| DDY903 (alias KKY38) | MATa his3Δ200 ura3-52 leu2-3,112 lys2-801am | D. Drubinc |
| DDY1102 (alias KKY49) | MATa/α his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 ura3-52/ura3-52 ADE2/ade2-1 lys2-801am/LYS2 | Kozminski et al. (2000) |
| HT195 | MATa his3-11 leu2-3,112 trp1-1 ura3-52 ade2-1 can1-100 bar1 CDC55-3xHA::his5 GAL+ | D. Kelloggd,e |
| JMY1172 | MATa cdc24-1 mih1::TRP1 bar1 ade1 his2 leu2-3,112 trp1 ura3Δns | D. Lewa,b |
| JMY1189 | MATa cdc24-1 mih1::TRP1 swe1::LEU2 bar1 ade1 his2 leu2-3,112 trp1 ura3Δns | D. Lewa,b |
| KKY164 | MATa/α zds2::kanMX4/ZDS2 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 ura3-52/ura3-52 ADE2/ade2-1 lys2-801am/LYS2 | This studyf |
| KKY168 | MATα zds2::kanMX4 his3Δ200 ura3-52 leu2-3,112 ade2-1 | Meiotic product of KKY164 |
| KKY169 | MATa zds2::kanMX4 his3Δ200 ura3-52 leu2-3,112 lys2-801am | Meiotic product of KKY164 |
| KKY542 | MATa zds1::HIS3 zds2::kanMX4 his3Δ200 ura3-52 leu2-3,112 lys2-801am | Meiotic product of KKY546 |
| KKY546 | MATa/α zds1::HIS3/ZDS1 ZDS2/zds2::kanMX4 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 ura3-52/ura3-52 ADE2/ade2-1 lys2-801am/LYS2 | KKY168 × KKY549 |
| KKY549 | MATa zds1::HIS3 his3Δ200 ura3-52 leu2-3,112 lys2-801am | Meiotic product of KKY1156 |
| KKY1075 | KKY1056 [YCplac111] | This studyf |
| KKY1076 | KKY1056 [pKK1575, YCplac111 (ZDS2)] | This studyf |
| KKY1077 | KKY542 [pKK1575, YCplac111 (ZDS2)] | This studyf |
| KKY1078 | KKY542 [YCplac111] | This studyf |
| KKY1081 | KKY542 [pKK1876, YCplac111 (zds2-3)] | This studyf |
| KKY1091 | MATa zds1::HIS3 zds2-i-9xmyc his3Δ200 ura3-52 leu2-3,112 lys2-801am | This studyf |
| KKY1101 | MATa zds1::HIS3 zds2::kanMX4 his3Δ200 ura3-52 leu2-3,112 lys2-801am [pKK1884, YCplac111 (zds2-i-9xmyc)] | This studyf |
| KKY1103 | MATa zds1::HIS3 zds2::kanMX4 his3Δ200 ura3-52 leu2-3,112 lys2-801am [pKK1886, YCplac111 (zds2-3-i-9xmyc)] | This studyf |
| KKY1124 | KKY542 [pKK1916, YCplac111 (zds2Δ129-479)] | This studyf |
| KKY1125 | KKY542 [pKK1920, YCplac111 (zds2Δ573-882)] | This studyf |
| KKY1126 | KKY542 [pKK1917, YCplac111 (zds2Δ13-130)] | This studyf |
| KKY1133 | KKY542 [pKK1918, YCplac111 (zds2Δ808-912)] | This studyf |
| KKY1127 | KKY542 [pKK1919, YCplac111 (zds2Δ813-942)] | This studyf |
| KKY1156 | MATa/α ZDS1/zds1::HIS3 zds2::kanMX4/ZDS2 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 ura3-52/ura3-52 ADE2/ade2-1 lys2-801am/LYS2 | This studyf |
| KKY1158 | KKY542 [M2739, YEp24 (ZDS1)] | This studyf |
| KKY1182 | MATa his3Δ200 ura3-52 leu2-3,112 lys2-801am cdc55::natMX | This studyf |
| KKY1183 | DLY657 [pDD42, pRS316 (GAL)] | This studyf |
| KKY1184 | DLY657 [pKK1930, pRS316 (GAL-ZDS1)] | This studyf |
| KKY1185 | DLY690 [pDD42, pRS316 (GAL)] | This studyf |
| KKY1186 | DLY657 [pKK1931, pRS316 (GAL-ZDS2)] | This studyf |
| KKY1187 | DLY657 [pKK1932, pRS316 (GAL-HSL7)] | This studyf |
| KKY1188 | JMY1172 [pDD42, pRS316 (GAL)] | This studyf |
| KKY1189 | JMY1172 [pKK1930, pRS316 (GAL-ZDSl)] | This studyf |
| KKY1190 | JMY1172 [pKK1931, pRS316 (GAL-ZDS2)] | This studyf |
| KKY1191 | JMY1172 [pKK1932, pRS316 (GAL-HSL7)] | This studyf |
| KKY1192 | JMY1189 [pDD42, pRS316 (GAL)] | This studyf |
| KKY1194 | MATa cdc24-1 swe1::LEU2 cdc55::kanMX4 bar1 ade1 his2 leu2-3,112 trp1 ura3Δns | This studyf |
| KKY1195 | MATa cdc24-1 cdc55::kanMX4 bar1 ade1 his2 leu2-3,112 trp1 ura3Δns | This studyf |
| KKY1200 | MATa cdc55::natMX zds1::HIS3 zds2::kanMX4 his3Δ200 ura3-52 leu2-3,112 lys2-801am | This studyf |
| KKY1201 | KKY1200 [pCMY18, pRS416 (CDC55)] | This studyf |
| KKY1203 | KKY1194 [pDD42, pRS316 (GAL)] | This studyf |
| KKY1204 | KKY1195 [pDD42, pRS316 (GAL)] | This studyf |
| KKY1205 | KKY1195 [pKK1930, pRS316 (GAL-ZDS1)] | This studyf |
| KKY1206 | KKY1195 [pKK1931, pRS316 (GAL-ZDS2)] | This studyf |
| KKY1207 | KKY1195 [pKK1932, pRS316 (GAL-HSL7)] | This studyf |
| KKY1217 | KKY1178 [pKK1954, pRS316 (GAL-zds2Δ808-912)] | This studyf |
| KKY1218 | MATa zds1::HIS3 zds2::kanMX4 rts1::natMX his3Δ200 ura3-52 leu2-3,112 lys2-801am | This studyf |
| PJ694a | MATa trp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ LYS2:GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. (1996) |
| PJ694α | MATα trp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ LYS2:GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. (1996) |
Strains are S288C unless indicated otherwise.
a BF264-15DU strain background.
b Gift of D. Lew (Duke University, Durham, North Carolina).
c Gift of D. Drubin (University of California, Berkeley).
d Gift of D. Kellogg (University of California, Santa Cruz).
e W303 strain background.
f See Materials and Methods.
Table 2.
Plasmids used in this study
| Plasmid | Relevant genotype | Source |
|---|---|---|
| M2654 | pTF63 (ZDS2) | Yu et al. (1996) |
| M2739 | YEp24 (ZDS1) | Yu et al. (1996) |
| p4339 | TA::MX4-natR switcher cassette | Tong et al. (2001) |
| pDD42 (alias pRB1438) | pRS316 (GAL) | Kozminski et al. (2000) |
| pKK1575 | YCplac111 (ZDS2) | This studya |
| pKK1802 | pAlter1 (zds1::HIS3) | Bi and Pringle (1996) |
| pKK1876 | Ycplac111 (zds2-3R863H, V868A) | This studya |
| pKK1884 | YCplac111 (zds2-i-9xmyc) | This studya |
| pKK1886 | YCplac111 (zds2-3-i-9xmyc) | This studya |
| pKK1916 | YCplac111 (zds2Δ129-479) | This studya |
| pKK1917 | YCplac111 (zds2Δ13-130) | This studya |
| pKK1918 | YCplac111 (zds2Δ808-912) | This studya |
| pKK1919 | YCplac111 (zds2Δ813-942) | This studya |
| pKK1920 | YCplac111 (zds2Δ573-882) | This studya |
| pKK1930 | pRS316 (GAL-ZDS1) | This studya |
| pKK1931 | pRS316 (GAL-ZDS2) | This studya |
| pKK1932 | pRS316 (GAL-HSL7) | This studya |
| pKK1934 | pGEX-4T1 (GST-ZDS2(731-942)) | This studya |
| pKK1938 | YCplac111(ZDS2) | This studya |
| pKK1939 | pOBD2 (GAL4DBD-ZDS2) | This studya |
| pKK1940 | pOBD2 (GAL4DBD-zds2Δ808-912) | This studya |
| pKK1941 | pRS425 (CDC55) | This studya |
| pKK1944 | pMAL-2c (CDC55) | This studya |
| pKK1954 | pRS316 (GAL- zds2Δ808-912) | This studya |
| pGEX-4T1 | (GST) | GE Healthcare |
| pMAL-2c | (MBP) | New England Biolabs |
| pML1 | pFA6a-kanMX6 | Longtine et al. (1998) |
| pOBD2 | CEN TRP1 GAL4-DBD | Uetz et al. (2000) |
| pOM22 | pOM (9xmyc:K. lactis URA3) | Gauss et al. (2005)b |
| pRS425 | 2μ LEU2 | Sikorski and Hieter (1989) |
| pSH65 | pGAL-cre | Gueldener et al. (2002) |
| pCMY14 | pRS424 (CDC55) | D. Burkec |
| pCMY18 | pRS416 (CDC55) | D. Burkec |
| YEplac195 | 2μ URA3 | Gietz and Sugino (1988) |
| YCplac33 | CEN URA3 | Gietz and Sugino (1988) |
| YCplac111 | CEN LEU2 | Gietz and Sugino (1988) |
All plasmids confer ampicillin resistance.
a See Materials and Methods.
b Obtained from EUROSCARF (Frankfurt, Germany).
c Gift of D. Burke (University of Virginia, Charlottesville, VA).
The method of Longtine et al. (1998) was used to delete ZDS2 in DDY1102, forming KKY164. PCR was used to amplify a kanMX4 disruption cassette with Bio-X-Act Long polymerase (Bioline, Tauton, MA) from template pML1 with primers oKK60 and oKK62. All primers used in this study are listed in Supplemental Table 1. Deletion of CDC55 in KKY38 was made by the same method, forming KKY1182. For this deletion, primers oKY33b and oKY34b were used to amplify a natMX disruption cassette from template p4339. Deletion of CDC55 was verified phenotypically (elongated buds at 18°C) and by PCR using diagnostic primers oKY35 and oKK135. Deletion of CDC55 in KKY542, which generated KKY1200 (cdc55Δ zds1Δ zds2Δ), was performed in the same manner. Rescue of the deletion was performed by transformation with pCMY18 (CDC55) forming KKY1201. Deletion of RTS1 in KKY542, which generated KKY1218 (rts1Δ zds1Δ zds2Δ), was performed in the same manner as the above CDC55 deletions, but using primers oKK247 and oKK248. Deletion was verified by PCR using primers oKK247 and oKY106.
The method of Longtine et al. (1998) was used to delete CDC55 in DLY657 and DLY690 using primers oKY100 and oKY101 to create KKY1195 (cdc24-1 cdc55Δ) and KKY1194 (cdc24-1 cdc55Δ swe1Δ). The deletion was verified by PCR using diagnostic primers oKY97, oKY99, and oKY49.
To delete ZDS1 in a zds2Δ/ZDS2 strain, an SmaI-SalI fragment from pKK1802, which contains HIS3 with ZDS1 flanking sequence, was transformed into KKY164, forming KKY1156.
Epitope Tagging of Zds2p
The method of Gauss et al. (2005) was used to introduce a 9xmyc epitope tag internal to the ZDS2 coding sequence in KKY549, forming KKY1091. In brief, primers oKY20 and oKY21 were used to amplify the 9xmyc:K.lactis URA3 cassette from pOM22. The PCR product was transformed into KKY549, which replaced codons 12–130 with the cassette. After integration, the strain was transformed with pSH65 (pGAL-Cre). Transformants were selected on YPD containing 50 mM potassium phosphate buffer, pH 7.0, and 50 μg/ml Zeocin (Invitrogen, Carlsbad, CA). Individual colonies were then picked and grown in liquid culture of the same selective medium. To induce Cre recombinase expression, cells were washed thrice with SC medium in which 2% glucose was substituted with 2% raffinose and 2% galactose (SC+Raf+Gal). After the final wash, cells were resuspended in 2 ml SC+Raf+Gal medium and grown 4 h. To recover strains in which Cre recombinase removed the URA3 marker from the ZDS2 coding region, the culture was serially diluted 1:2000 with SC medium, of which 200 μl was plated on SC medium. After 1-d growth, colonies were replica plated on SC-Ura and SC medium. After 2-d growth, colonies that grew on SC, but not SC-Ura were picked and struck onto SC plates containing 0.5 mg/ml 5-fluoroorotic acid (5-FOA). Strains surviving on SC+FOA medium, but not SC-Ura were picked for further analysis. DNA sequencing verified the tag was in-frame.
Plasmids
To construct a CEN plasmid containing ZDS2 (pKK1575), ZDS2 was amplified from plasmid template M2654 with Bio-X-Act Long polymerase using primers oKK148 and oKK149 and subcloned into the SacI and BamHI sites of YCplac111. DNA sequencing confirmed the fidelity of amplification. In comparison to the Saccharomyces Genome Database (SGD) Zds2p sequence, pKK1575 encodes a Zds2p with amino acid substitutions M7V, T23S, A530P, A546G, N568S, and A729E; deletion of T723; and an insertion of residues NC between V711 and E712. Amino acids in this report are numbered according to the SGD reference sequence. Functionality of amplified ZDS2 was determined by the ability of pKK1575 to rescue the bud growth defect of KKY542 (zds1Δ zds2Δ).
CEN plasmids encoding Zds2p or Zds2–3p with a 9xmyc epitope tag, pKK1884 and pKK1886, respectively, were constructed by gap repair. 9xmyc flanked by ZDS2 sequence was amplified from KKY1091 using primers oKY31 and oKY32. This PCR product was then cotransformed into KKY169 with pKK1575 (ZDS2) or pKK1876 (zds2-3) gapped with SwaI and StuI. From transformants selected on SC-Leu plates, gap repaired plasmids were isolated and sequenced with primer oKK205.
To construct a 2μ-plasmid with CDC55 (pKK1941), a XhoI-NotI fragment containing CDC55 was subcloned from pCMY14 into the XhoI-NotI sites of pRS425. Functionality of the construct was confirmed by the rescue of the cdc55Δ cold-sensitive elongated bud phenotype in strain KKY1182.
Microscopy
Epifluorescence microscopy was performed with a Nikon E800 microscope equipped with a 100x/1.3 Plan Fluor objective (Nikon, Melville, NY). Images were captured with an Orca 100ER digital camera (Hamamatsu Photonics, Hamamatsu City, Japan) and Openlab software (Improvision, Lexington, MA). Contrast enhancement was linear for each image series. Preparation of cells for indirect immunofluorescence was performed as in Kozminski et al. (2006) unless otherwise indicated. Mouse anti-c-myc/9E10 (Santa Cruz Biotechnology, Santa Cruz, CA) and Alexa 568–conjugated goat anti-mouse (Molecular Probes/Invitrogen) antibodies were diluted 1:100 in PBS (pH 7.4) containing 1 mg/ml BSA.
Differential interference contrast (DIC) microscopy was performed using a Nikon E800 microscope equipped with a 100×, 1.4 NA Plan Apo objective. Images were captured and processed as described above, with background subtraction. Cells were prepared for DIC microscopy by gently pelleting log-phase cultures, resuspending them in SC medium lacking glucose, and placing 3.5 μl of the cell suspension between an acid-washed slide and coverslip.
Screen for Temperature-conditional zds2 Alleles
Fifty cycles of degenerate PCR were used to amplify independent pools of the ZDS2 coding sequence between codons 821 and 906. Each 100 μl PCR reaction contained Thermophilic DNA Polymerase Buffer (Promega, Madison WI), 1.5 mM MgCl2, 130 μM MnCl2, 200 μM dCTP, 200 μM dTTP, 40 μM dATP, 40 μM dGTP, 0.4 μM primers (oKK210 and oMR1), 100 pg pKK1575 template, and 0.8 μl Taq polymerase [5 U/μL] (Promega). To introduce the mutagenized coding sequence into ZDS2 by gap repair, each pool of PCR product was cotransformed into KKY1158 with YCplac111 (ZDS2; pKK1575) linearized with SmaI. Transformants were selected on SC-Leu plates at a density of ∼200 colonies per plate. Because KKY1158 contained YEp24 (ZDS1) to rescue the zds1Δ zds2Δ growth defect, a plasmid shuffle was then used to select transformants containing YCplac111 (zds2) but not YEp24 (ZDS1). Thus, every transformant subsequently analyzed depended only on YCplac111 (zds2) to rescue the zds1Δ zds2Δ growth defect. After 4–5 d growth on SC-Leu plates, transformants were replica plated onto SC-Leu plates containing 0.75 g/l FOA (US Biological, Swampscott, ME), which counterselected YEp24 (ZDS1). Of two replicas for each transformation plate, one was incubated for 2–3 d at 25°C, whereas the other was incubated for 2–3 d at 37°C. Colonies that grew at 25°C but exhibited little or no growth at 37°C were designated putative temperature-sensitive mutants. These putative mutants were picked from the 25°C replica plate and restreaked onto SC-Leu plates. From each of the clonal purifications, 2–3 individual colonies were picked for further analyses. In total, ∼10,000 colonies were screened. To determine whether temperature-sensitive growth was dependent upon a mutation in ZDS2, plasmids were isolated from each putative temperature-sensitive mutant and retransformed into KKY542. Plasmids that conferred temperature-sensitive growth or morphology defects at 37C but not 25°C, either on SC-Leu plates (5 d) or in SC-Leu asynchronous liquid culture, were then sequenced using primers oKK210 and oMR1. ZDS2 in pKK1575 was used as the reference sequence.
Structure–Function Mapping of Zds2p
To map conserved regions of Zds2p necessary for regulating bud growth, a series of plasmids were made in which Zds2p was truncated or contained an in-frame deletion within the ZDS2 coding sequence. These constructs were then transformed individually into KKY542 (zds1Δ zds2Δ) and the bud morphology of the resulting transformants was analyzed by DIC microscopy.
The plasmids used to map the structure-function of Zds2p are the following. pKK1916 (zds2Δ129-479), which contains an in-frame deletion of residues 129-479, was constructed by digesting pKK1575 (ZDS2) with NsiI and ligating the resulting 8.8-kb fragment. pKK1917 (zds2Δ13-130), which contains an in-frame deletion of residues 13-130, was constructed by digesting pKK1575 with MscI and NdeI (blunted with Mung Bean nuclease) and ligating the 9.5-kb fragment. Construction of this plasmid introduced a silent C-to-T mutation at base pairs + 36/codon 12. pKK1920 (zds2Δ573-882), containing an in-frame deletion of residues 573-882, was constructed by digesting pKK1575 with HpaI and SmaI and ligating the 8.8-kb fragment. pKK1918 (zds2Δ808-912), which contains an in-frame deletion of residues 808-912, was constructed in two steps by using the Quick Change XL Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA). First, an NruI site was added to pKK1575 at base pairs +2424 using primers oKY8 and oKY9 to replace the A at base pairs +2424 with GCGA. Furthermore a BsaAI site was added by adding bases CAC after base pairs +2733. The plasmid produced, pKK1938, was then digested with NruI and BsaAI. The resulting 9.5-kb fragment was then ligated to create pKK1918 (zds2Δ808-912). (Construction of this plasmid introduced a silent A to G mutation at base pairs + 2424/codon 808). DNA sequencing verified the coding sequence remained in-frame. pKK1919 (zds2Δ813-942), which contains an Stop codon after residue 812, was constructed using pKK1575 and the Quick Change XL Site-directed Mutagenesis Kit (Stratagene) with primers oKY6 and oKY7c, which introduced a C-to-T substitution at base pairs +2437 (Stop) and an A-to-G substitution at base pairs +2440 (created AflII site for diagnostic digest). Stop codon insertion was verified by DNA sequencing.
Differential Two-Hybrid Screen for Zds2p-interacting Proteins
To identify proteins that require the ZH4 domain for binding to Zds2p, two-hybrid assays were performed with both wild-type Zds2p and Zds2p lacking the ZH4 domain (Zds2Δ808-912p). Two-hybrid expression plasmids were created as follows. ZDS2 was PCR amplified using primers oKY12 and oKY13 and template pKK1575. zds2Δ808-912 was amplified with the same primers from template pKK1918. The ZDS2 and zds2Δ808-912 amplicons were each digested with NcoI and PstI and cloned into the NcoI/Pst1sites of yeast vector pOBD2 by ligation with T4 DNA ligase. The inserts are in-frame with the coding sequence for the Gal4p DNA binding domain (Gal4p-DBD), forming pKK1939 and pKK1940, respectively. Clones were verified by DNA sequencing and transformed into yeast strain PJ69-4α (MATα). Two individual colonies of each construct were mated to an ordered array containing S. cerevisiae ORFs fused to the Gal4p activation domain (Gal4p-AD; Yeast Resource Center, University of Washington, Seattle, WA) in yeast strain PJ69–4a (MATa), as described by Uetz et al. (2000). Diploid yeast resulting from these matings were replica-plated onto selective media (SC-Trp-Leu-His + 3 mM 3-amino-1,2,4-triazole [3AT] and incubated at 30°C for at least 1 wk before being scored for growth).
Putative interactors of Zds2p identified in this genome-wide screen as well as those previously reported in the literature (Drees et al., 2001) were then chosen for small-scale assays. Yeast strains that expressed these Gal4p-AD-yeast ORF fusions were selected from the array and mated with strains that contained full-length and mutant (Zds2Δ808-912p) fused to Gal4p-DBD. Strains that contained constructs for Gal4p-DBD, Gal4p-AD, Gal4p-DBD-Rad17p, and Gal4p-AD-Mec3p (Rad17p and Mec3p are known to interact) were included as controls. Mating reactions were serially diluted and spotted onto medium that selects for diploids (SC-Trp-Leu) or for interactors (SC-Trp-Leu-His + 3AT) and scored for growth as described above.
Assay for Progression through the Swe1p-dependent G2/M Checkpoint
To analyze the regulation of the Swe1p-dependent G2/M checkpoint, several CEN plasmids were created that put ZDS1, ZDS2, zds2Δ808-912, and HSL7 under regulation of a GAL1/10 promoter. pKK1930 (GAL-ZDS1) was constructed by PCR amplification of ZDS1 from plasmid template M2739 with primers oKY54 and oKY55, which add a BamHI site directly upstream of the Start codon and an SpeI site 42 base pairs (bp) downstream of the Stop codon, respectively. The PCR product was digested with BamHI and SpeI and cloned into pDD42 (GAL1/10) digested with the same enzymes. pKK1931 (GAL-ZDS2) and pKK1954 (GAL-zds2Δ808-912) were constructed by PCR amplification of ZDS2 or zds2Δ808-912, respectively, from templates pKK1575 (ZDS2) or pKK1918 (zds2Δ808-912) with primers oKY77 and oKY50, which added a BamHI site directly upstream of the Start codons. The PCR products were digested with BamHI and SacI (a site 266 bp downstream of the Stop codon in the templates) and subcloned into pDD42. pKK1932 (GAL-HSL7) was made by PCR amplification of HSL7 from a genomic DNA preparation of KKY38 with primers oKY56 and oK76 which added a BamHI site directly upstream of the Start codon and an SacI site 466 bp downstream of the Stop codon, respectively. This PCR product was then digested with BamHI and SacI and subcloned into pDD42 digested with the same enzymes.
To determine whether ZDS1, ZDS2, zds2Δ808-912, or HSL7 negatively or positively regulate the Swe1p-dependent G2/M checkpoint, plasmids containing GAL-ZDS1 (pKK1930), GAL-ZDS2 (pKK1931), GAL-zds2Δ808-912 (pKK1954), GAL-HSL7 (pKK1932), or an empty GAL vector (pDD42) were transformed into a strain (DLY657) containing cdc24-1, which confers an actin organization defect at 37°C, generating strains KKY1183 (cdc24-1 [GAL]), KKY1184 (cdc24-1[GAL-ZDS1]), KKY1186 (cdc24-1 [GAL-ZDS2]), KKY1217 (cdc24-1 [GAL zds2Δ808-912]), and KKY1187 (cdc24-1[GAL-HSL7]). Strain KKY1185 (swe1Δ cdc24-1[GAL]) was constructed by transforming DLY690 with pDD42. Strains were grown overnight at 25°C in a baffled flask with vigorous shaking in complete synthetic medium lacking uracil (CSM-Ura; 0.69 g/l complete synthetic medium-Ura supplement [MP Biomedicals, Solon, OH], 5 g/l ammonium sulfate ([Sigma-Aldrich, St. Louis, MO], 1.5 g/l Difco yeast nitrogen base without amino acids and ammonium sulfate ([BD Biosciences, Franklin Lakes, NJ]) and containing 2% raffinose and 2% galactose in place of glucose. The following morning, log-phase cells were harvested by centrifugation and resuspended in YP medium (no glucose) containing 2% raffinose and 2% galactose (YP+Raf+Gal) and then arrested in G1 with 0.75 μg/ml α-factor (synthesized at the University of Virginia Biomolecular Core Facility) for 2.5 h at 25°C. Phase-contrast microscopy verified that >90% of the cells in each culture were unbudded, indicative of G1 arrest. Strains were released from α-factor arrest by washing twice with YP+Raf+Gal medium prewarmed to 37°C. Three-milliliter aliquots were fixed at regular intervals with 4.75% (vol/vol) formaldehyde. Fixed cells were then washed with PBS and permeabilized with 0.1% Triton X-100 for 4 min. After two washes with PBS to remove the detergent, cells were resuspended in 1 ml PBS with 0.12 μg/ml DAPI (4′,6-diamidino-2-phenylindole). Unbudded cells were then scored by fluorescence microscopy as mononucleate or multinucleate. To verify that recombination did not occur between plasmid borne GAL-zds2Δ808-912 and endogenous ZDS1 or ZDS2, we sequenced plasmid rescued from KKY1217 after the assay.
To determine whether ZDS1, ZDS2, or HSL7 negatively or positively regulate the Swe1p-dependent G2/M checkpoint in the absence of CDC55, plasmids containing GAL-ZDS1 (pKK1930), GAL-ZDS2 (pKK1931), GAL-HSL7 (pKK1932), or an empty GAL vector (pDD42) were transformed into a strain (KKY1195) containing cdc24-1 cdc55Δ resulting in strains KKY1205 (cdc24-1 cdc55Δ [GAL-ZDS1]), KKY1206 (cdc24-1 cdc55Δ [GAL-ZDS2]), KKY1207 (cdc24-1 cdc55Δ [GAL-HSL7]) and KKY1204 (cdc24-1 cdc55Δ [GAL]). Strain KKY1203 (swe1Δ cdc55Δ cdc24-1 [GAL]) was constructed by transforming KKY1194 with pDD42. Cells were grown at 30°C and assayed at 37°C as described above.
To determine whether ZDS1, ZDS2, or HSL7 negatively or positively regulate the Swe1p-dependent G2/M checkpoint in the absence of MIH1, plasmids containing GAL-ZDS1 (pKK1930), GAL-ZDS2 (pKK1931), GAL-HSL7 (pKK1932), or an empty GAL vector (pDD42) were transformed into a strain (JMY1172) containing cdc24-1 mih1Δ, resulting in strains KKY1189 (cdc24-1 mih1Δ [GAL-ZDS1]), KKY1190 (cdc24-1 mih1Δ [GAL-ZDS2]), KKY1191 (cdc24-1 mih1Δ [GAL-HSL7]) and KKY1188 (cdc24-1 mih1Δ [GAL]). Strain KKY1192 (swe1Δ mih1Δ cdc24-1 [GAL]) was constructed by transforming JMY1189 with pDD42. Cells were grown at 25°C and assayed at 37°C as described above.
SDS-PAGE and Immunoblots
Unless otherwise indicated, gels were prepared for SDS-PAGE as previously described (Kozminski et al., 2000) and electrotransferred to 0.2-μm nitrocellulose membranes (Bio-Rad, Hercules, CA) for 4 h at 75 V in using a Mini Trans-Blot Cell apparatus (Bio-Rad). Blots were blocked with 5% (wt/vol) Carnation instant milk (Nestle, Glendale, CA) in TBS for 30 min. Blots were washed thrice with TBS containing 0.1% (vol/vol) Tween 20 (Sigma-Aldrich) for 15 min after each antibody incubation. Primary antibodies, mouse anti-c-myc (9E10; Santa Cruz Biotechnology), rabbit anti-GST (Dighe and Kozminski, 2008), rabbit anti-myelin basic protein (MBP), or mouse anti-hemagglutinin (HA; 16B12; Covance, Princeton, NJ), was used at room temperature overnight at 1:1000 dilution; secondary antibody, horseradish peroxidase (HRP) anti-mouse (Amersham, Uppsala, Sweden), was used at room temperature for 30 min at 1:5000 dilution. Blots were incubated with SuperSignal West-Pico Chemiluminescent Substrate (Pierce, Rockford, IL) for 5 min and then exposed to X-OMAT Blue (Eastman Kodak, Rochester, NY) film.
Binding Assay with Cell Lysates
To determine whether the C-terminus of Zds2p is sufficient for interaction with Cdc55p, 3xHA-tagged Cdc55p was isolated from whole cell lysates using GST-Zds2(731-942)p–bound beads. To create a fusion of glutathione S-transferase (GST) with a C-terminal fragment of Zds2p (pKK1934), amino acids 731-942, the coding sequence of this fragment was amplified from pKK1575 by PCR using oligonucleotides oKY83 (puts a BamHI site 5′ of codon 731) and oKY87 (puts an SalI site 3′ of the Stop codon). This fragment was then digested with BamHI/SalI and subcloned into pGEX-4T1 (GE Healthcare, Piscataway, NJ) digested with the same enzymes. DNA sequencing verified an in-frame fusion.
To express GST and GST-Zds2(731-942)p, pGEX-4T1 and pKK1934 were transformed individually into Escherichia coli strain BL21(DE3). Cultures of 300 ml were grown to 0.6–0.8 O.D.600 nm in LB medium with 100 μg/ml ampicillin, before supplementation with 2% ethanol and induction with 1 mM IPTG for 3 h at 37°C. Bacteria were then pelleted and stored at −80°C until lysis.
To prepare GST-Zds2(731-942)p bound glutathione-Sepharose beads, bacteria were resuspended in 10 ml bacterial lysis buffer (BLB; 50 mM Tris, pH 8.0, 20% [wt/vol] sucrose, 10% (vol/vol) glycerol, 2 mM DTT, 1 mM PMSF, and 0.5 μg/ml each of leupeptin, aprotinin, pepstatin A, chymostatin, and antipain) and lysed by sonication at 4°C. The lysate was centrifuged at 13,200 × g for 20 min at 4°C, and the supernatant was decanted onto 125 μl glutathione-Sepharose beads (GE Healthcare), equilibrated with 10 ml BLB. Beads and supernatant were nutated for 1 h at 4°C. The beads were then washed with 15 mL BLB 6 times at 4°C. The amount and homogeneity of the GST proteins bound to the beads were visually estimated on a Coomassie Blue–stained SDS-PAGE gel, with known masses of BSA serving as standards.
To pull down Cdc55p-3xHA from yeast cell lysates, glutathione-Sepharose beads containing either 2 μg GST or GST-Zds2(731-942)p were preequilibrated with yeast lysis buffer (50 mM HEPES-KOH, pH 7.6, 100 mM β-glycerolphosphate, 25 mM NaF, 1 mM MgCl2, 1 mM EGTA, 5% glycerol, 0.15% Tween-20, 1 mM PMSF, 150 mM NaCl, and 0.5 μg/ml each of leupeptin, aprotinin, pepstatin A, chymostatin, and antipain) and then added to 400-μl aliquots of clarified yeast cell lysate. The beads and lysate were nutated for 4 h at 4°C. Beads were then washed with 1 mL yeast lysis buffer 5 times, resuspended in 100 μl of 2× sample buffer (2% SDS, 10% glycerol, 0.1 M Tris-Cl, pH 7.6, 2 mM EDTA, 80 mM DTT, and 0.1 mg/ml bromophenol blue), and analyzed by SDS-PAGE and immunoblotting. To prepare the yeast lysate, 0.23 g of yeast powder were thawed in 1 ml yeast lysis buffer and clarified for 5 min at 14,000 × g at 4°C. The yeast powder was made from log-phase HT195 cells, harvested by centrifugation, and lysed with a pestle in a mortar filled with liquid nitrogen. Yeast powder was stored at −80°C until use.
Binding Assay with Recombinant Proteins
To test whether the ZH4 domain of Zds2p binds directly to Cdc55p, MBP-Cdc55p and GST-Zds2(731-942)p were expressed in E. coli, purified, and mixed in vitro. GST-Zds2(731-942)p and GST were expressed and purified as described above, except they were eluted from the glutathione-Sepharose beads with 10 mM reduced glutathione, followed by dialysis in PBS. MBP-Cdc55p was expressed from pKK1944 and purified in the same manner as GST-Zds2(731-942)p, except that 0.125 mM IPTG was used for induction and the clarified lysate was mixed with 125 μl amylose resin (New England Biolabs, Ipswich, MA) preequilibrated with 10 ml BLB. Maltose at 10 mM in BLB was used to elute MBP-Cdc55p from the resin. To construct pKK1944 the coding sequence of CDC55 was amplified by PCR, using the template pCMY14 and primers oKK229 and oKK230, and subcloned into the HindIII and XmnI sites of pMAL-2c. Binding of MBP-Cdc55p to either GST or GST-Zds2(731-942)p was tested by incubating 0.4 mM MBP-Cdc55p and 10 μl of equilibrated glutathione-Sepharose resin with 0.4 mM GST or GST-Zds2(731-942)p in binding buffer (50 mM HEPES-KOH, pH 7.6, 1 mM MgCl2, 1 mM EGTA, 5% glycerol, 0.15% Tween-20, 150 mM NaCl) in a 50 μl total reaction volume. The reaction was incubated for 30 min at 4°C, and then the beads were spun down and the supernatant saved as the unbound fraction for further analysis. The beads were then washed with 1 mL of wash buffer 6 times (50 mM HEPES-KOH, pH 7.6, 1 mM MgCl2, 1 mM EGTA, 5% glycerol, 0.15% Tween 20, 500 mM NaCl, 2 M urea) before resuspending beads (bound sample) in 2× protein sample buffer.
RESULTS
Structure–Function Mapping of Zds2p Revealed a Domain That Regulates Bud Morphology
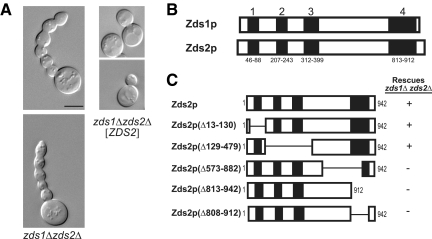
The suppression of the elongated bud phenotype of zds1Δ zds2Δ cells by the loss of Swe1p kinase (McMillan et al., 1999b) strongly suggests Zds proteins regulate the Swe1p-dependent G2/M checkpoint shown in Figure 1. As an initial step toward testing this idea and defining the mechanism of regulation, we asked what regions of Zds2p are necessary for regulating bud morphology, which we used as a read-out of cell cycle progression (Hartwell et al., 1974). Because either plasmid-borne ZDS2 (Figure 2A, right) or ZDS1 (data not shown) rescued the elongated bud morphology of zds1Δ zds2Δ cells (Figure 2A, left), we hypothesized that Zds proteins contain a conserved functional domain. Using a Clustal W2 amino acid sequence alignment algorithm (Larkin et al., 2007), we identified four regions of high homology (>75% sequence identity) between Zds1p and Zds2p (Figure 2B). To test whether any of these Zds homology (ZH) regions are necessary for the regulation of bud morphology, we transformed a zds1Δ zds2Δ haploid strain with a CEN plasmid that contained either wild-type ZDS2 or a mutant allele containing an in-frame deletion of one or more ZH coding regions. The resultant transformants were analyzed by DIC microscopy to determine whether the elongated bud morphology remained. We found that large regions of Zds2p are dispensable for bud growth regulation (Figure 2C). In contrast, a C-terminal region consisting of amino acids 813-912, which we refer to as ZH4, was necessary for the rescue of the elongated bud phenotype of zds1Δ zds2Δ cells. Identification of ZH4 as an important functional domain is consistent with previous work by Schwer et al. (1998) that showed expression of, approximately, the last 150 amino acids of the C-terminus of paralogs Zds1p or Zds2p is sufficient to rescue the mutant bud morphology of a zds1Δ zds2Δ strain. Thus, the ZH4 domain is essential for regulating bud growth and likely cell cycle progression as well.
Figure 2.
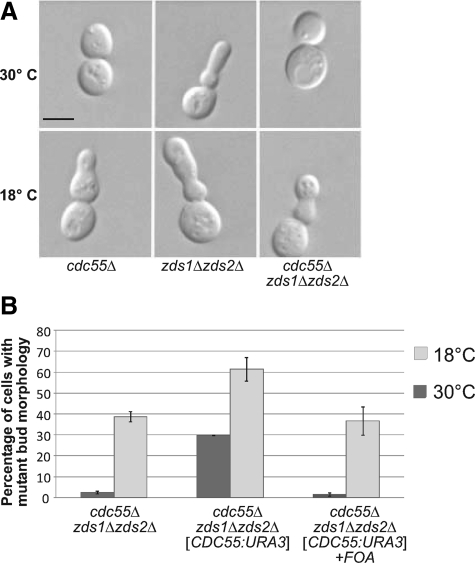
Structure-function mapping of Zds2p shows the ZH4 domain in necessary for the regulation of bud growth. (A) DIC micrographs of isogenic log-phase S. cerevisiae zds1Δ zds2Δ strains, grown in SC-Leu medium at 25°C, transformed with a CEN vector (KKY1078) or the same vector containing ZDS2 (KKY1077). Scale bar, 5 μm. zds1Δ zds2Δ strains produce elongated buds with multiple constrictions along the length of the bud. “Elongated bud” is defined as a daughter bud that has an axial length longer than the mother cell. (B) Four regions of high amino acid identity (>75%) between Zds1p and Zds2p as identified with a Clustal W alignment. (C) Wild-type and mutant Zds2p constructs used for Zds2p structure–function mapping. CEN plasmids encoding these constructs were transformed into a zds1Δ zds2Δ strain (KKY542) and scored by DIC microscopy for rescue of zds1Δ zds2Δ bud morphology defects. Only constructs that contained the entire ZH4 region (amino acids 813-912) rescued zds1Δ zds2Δ bud morphology defects.
Mutagenesis of ZH4 Domain Resulted in Temperature-sensitive Bud Morphology and Zds2p Localization Defects
As further evidence for the functional relevance of the ZH4 domain we identified temperature-sensitive ZDS2 alleles via random mutagenesis of the domain. Using a degenerate PCR approach, we introduced random mutations in ZDS2 between codons 821 and 906 and screened, on the basis of colony size in a zds1Δ strain background, for alleles that confer a growth defect at 37°C. zds1Δ zds2Δ cells grow more slowly than wild-type and zds1Δ cells; therefore, mutations that affect ZDS2 function were predicted to confer a significant growth defect in combination with zds1Δ. One allele identified in the screen, zds2-3R863H,V868A, had a temperature-dependent bud morphology phenotype. At 25°C, zds1Δ zds2-3 cells formed buds with a wild-type morphology. When an asynchronous log phase culture was shifted to 37°C for 4 h, 42% of the budded zds1Δ zds2-3 cells (n = 229) exhibited elongated buds similar to those displayed by zds1Δ zds2Δ cells (Figure 3A). Bud morphology of zds1Δ ZDS2 cells remained unaffected by the shift to 37°C (Figure 3A). The zds2-3 allele is recessive because it did not confer a mutant phenotype if a wild-type copy of ZDS2 was present (data not shown). An effect on bud morphology was only observed when the allele was transformed into a zds1Δ zds2Δ strain (Figure 3A, right).
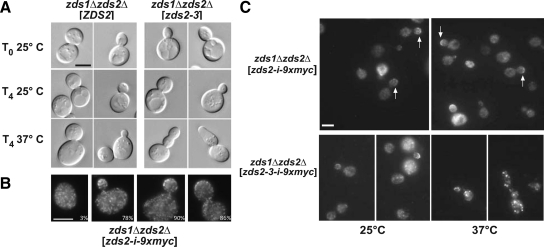
Figure 3.
A temperature-sensitive zds2 allele demonstrates the functional significance of ZH4. (A) DIC micrographs of log-phase zds1Δ zds2Δ strain transformed with a CEN plasmid that contained either a wild-type (ZDS2; KKY1076) or temperature-sensitive allele of ZDS2 (zds2-3ts; KKY1081). Cultures were incubated at 25°C (T0) in SC-LEU medium and then split for an additional 4 h of growth (T4) at 25 or 37°C. Note the elongated bud morphology of the zds2-3ts strain after shift to 37°C. (B) Zds2p localizes to the bud cortex and to cytoplasmic puncta. Indirect immunofluorescence micrographs show (left to right) the localization of Zds2p with an internal myc epitope tag (i-9xmyc) in representative log-phase unbudded, small-budded, medium-budded, and large-budded zds1Δ zds2-i-9xmyc cells (KKY1091) grown at 25°C in SC-LEU medium. The percentage of cells that showed myc-tagged Zds2p localization at the bud cortex or cell cortex in unbudded cells is given in the bottom right of each panel. n = 200 cells scored for each bud size category. (C) Bud cortex localization of Zds2p is lost in zds2-3ts cells at restrictive temperature. Indirect immunofluorescence micrographs of a log-phase isogenic zds1Δ zds2Δ strain transformed with either a CEN plasmid containing zds2-i-9xmyc (KKY1101) or zds2-3-i-9xmyc (KKY1103). Strains were grown at 25°C in SC-LEU and then split for growth at 25 and 37°C for 4 h. Scale bar, 5 μm.
To observe the localization of Zds2–3p, as well as wild-type Zds2p, we replaced amino acids 12-130 in these proteins with a 9x-myc tag. The amino acids replaced by the epitope tag were shown previously to be nonessential for the regulation of bud growth (Figure 2). An internal epitope tag was chosen because we were unable to detect wild-type Zds2p in cells, by fluorescence microscopy, when it was tagged at either its N- or C-terminus. Our internally tagged construct, zds2-i-9x-myc, rescued the mutant bud morphology of the zds1Δ zds2Δ strain and was found to localize at the bud cortex (Figure 3, B and C, white arrows in top panels) and as small puncta throughout the cell body (Figure 3B). In unbudded cells only 3% of the cells showed cortical localization of the tagged Zds2p. On budding, Zds2-i-9x-myc concentrated at the bud cortex in 78% of small-budded, 90% of medium-budded, and 86% of large-budded cells (n = 200 for each bud category). Interestingly, the product of zds2-3, Zds2–3p, is no longer observed on the bud cortex after the strain is shifted to a nonpermissive temperature. zds2-3-i-9x-myc rescued the mutant bud phenotype of the zds1Δ zds2Δ strain at 25°C (Figure 3C, bottom left panels) but, when the culture was shifted from 25°C to 37°C for 4 h, we observed a loss of the mutant protein from the bud tip and an accumulation of large fluorescent puncta in the cell body (Figure 3C, bottom right panels).
zds2-3 encodes two amino acid substitutions R863H and V868A. Although we have not tested whether either mutation alone confers a temperature-sensitive cell growth and bud morphology defect, it is notable that R863 is highly conserved (Supplemental Figure 1) in fungi of agricultural, pathogenic, and research interest, suggesting that it is important for Zds protein function across species. Based on sequence there are no obvious homologues of the Zds proteins in mammalian cells; however, some weak identity to Pus (pseudouridine synthase) 10-family proteins exists (Supplemental Figure 2).
ZH4 Is Necessary for Interaction with the PP2A Regulatory Subunit Cdc55p
To understand the molecular function of the ZH4 domain we sought to identify proteins that require this domain for interaction with Zds2p. To this end, we performed a genome-wide differential two-hybrid screen for proteins that bind to Zds2p but not to a Zds2p construct lacking the ZH4 domain. Baits, which were Gal4-DBD fused to either Zds2p or Zds2pΔ808-912, were screened against a Gal4-AD library. This genome wide screen returned one polypeptide, Cdc55p, which interacted with full-length Zds2p (Figure 4A, right panel) but not the mutant Zds2p lacking the ZH4 domain (Figure 4A, right panel). Protein kinase C (Pkc1p) was also found, but it interacted with both wild-type and mutant Zds2p (Figure 4A, right panels). With the exception of Pkc1p, for reasons unknown, none of the other reported (Drees et al., 2001) two-hybrid interactions with full-length Zds2p were found in this screen. Because Cdc55p is a component of PP2A, we performed a second screen with a reduced set of Gal4-AD fusions to determine whether Zds2p interacts with any other subunits of the phosphatase. We found that full-length Zds2p interacted with the structural (Tpd3p) and catalytic (Pph21p) subunits of PP2A but not the other regulatory subunit (Rts1p) present in S. cerevisiae (Figure 4A, right panels). Rad17p and Mec3p are known to interact (Lydall et al., 1996; Majka and Burgers, 2003; Shinohara et al., 2003) and served as a positive control for the assay. All interactions with Zds2p, except the interaction with Pkc1p, depended on the ZH4 domain. These data strongly suggested that ZH4 is a binding domain for PP2A(Cdc55p).
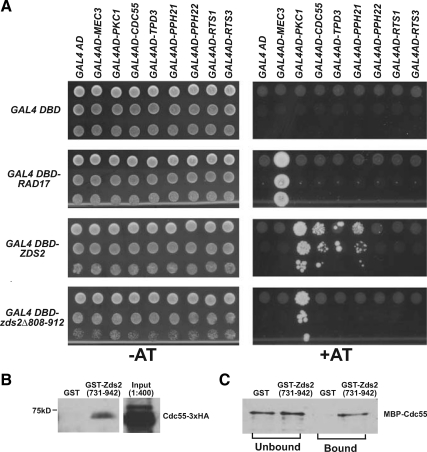
Figure 4.
Cdc55p binding to Zds2p requires the ZH4 domain. (A) A two-hybrid assay that showed Zds2p interacts with kinase Pkc1p and PP2A subunits Cdc55p (regulatory B subunit), Pph21p (catalytic subunit), and Tpd3p (structural subunit). The PP2A subunits, however, did not interact with Zds2Δ808-912p, which lacks the ZH4 domain. Neither construct interacted with Pph22p (catalytic PP2A subunit), Rts1p (PP2A regulatory B′ subunit), or Rts3p (Putative component of PP2A). Rad17p (DNA damage checkpoint protein) and Mec3p (DNA damage checkpoint protein) are known to interact. The Gal4-AD and Gal4-DBD controls for nonspecific colony growth. Strains were plated as 10-fold serial dilutions (top to bottom) onto selective media (SC-Trp-Leu-His) with (+AT) or without (−AT) 3 mM 3-amino-1,2,4-triazole (3AT) and incubated at 30°C for at least 1 wk before being scored for growth. Growth on 3AT indicates a protein–protein interaction. (B) GST-Zds2(731-942)p pulls down Cdc55-3HA from yeast lysate. GST (pGEX-4T1) or GST-Zds2(731-942)p bound to glutathione beads was incubated at 4°C for 4 h with the Cdc55-3HA containing lysate (KKY1107). One-fourth of the sample that bound to the beads was loaded, and the immunoblot probed with anti-HA antibody. Input represents 1:400 of the total lysate added to the beads. (C) MBP-Cdc55p binds GST-Zds2(731-942)p but not GST in an in vitro–binding assay. One-fifth of unbound and bound fractions were separated by SDS-PAGE, and immunoblots were probed with anti-MBP antibody.
To demonstrate that the C-terminal domain of Zds2p is sufficient for association with Cdc55p, we tested whether the C-terminal domain of Zds2p could affinity purify Cdc55p from yeast cell lysates. For this experiment, recombinant GST or GST-Zds2(731-942)p bound to glutathione-Sepharose beads were incubated with a cell lysate prepared from a yeast strain expressing Cdc55p-3xHA. We found that Cdc55p interacted with a fusion construct containing the ZH4 domain of Zds2p but not with GST alone (Figure 4B). Lysates lacking Cdc55p-3xHA showed no immunoreactivity with the anti-HA antibody (data not shown). Thus, the C-terminal domain of Zds2p is sufficient for association with Cdc55p.
To determine whether the C-terminal domain of Zds2p binds Cdc55p directly, we performed an in vitro binding assay with recombinant MBP-Cdc55p and an equimolar amount of either GST or GST-Zds2(731-942)p. We found that Cdc55p interacted with GST-Zds2(731-942)p but not GST alone (Figure 4C). This result showed that the C-terminal domain of Zds2p, which includes ZH4, binds to Cdc55p directly, as predicted by two-hybrid analysis.
Misregulation of Bud Morphology in a zds1Δ zds2Δ Strain Is Dependent on CDC55
In vivo, Cdc55p is present at a high stochiometric ratio with the Zds proteins (Ghaemmaghami et al., 2003). To demonstrate that the Cdc55-Zds2 protein interaction is physiologically relevant, we analyzed the effect of deleting these genes in combination. Deletion of CDC55 is known to cause a cold-sensitive phenotype that involves a mis-regulation of bud growth (Healy et al., 1991; Figure 5A, left panel). This phenotype is similar to the elongated bud morphology seen in the zds1Δ zds2Δ strain at all temperatures (cf. Figure 5A, left and middle panels). We found that deleting CDC55 in conjunction with the ZDS1 and ZDS2 paralogs rescued the elongated bud morphology of the zds1Δ zds2Δ strain at 30°C (Figure 5A, right panel). The same phenotype was observed with independently generated cdc55 Δ zds1Δ zds2Δ strains. Introduction of a CDC55-containing plasmid into a triple mutant strain (zds1Δ zds2Δ cdc55Δ) returned the bud morphology to a mutant, elongated state (Figure 5B). Thus, the presence of CDC55 in the zds1Δ zds2Δ strain is necessary for the mutant bud morphology, at least at 30°C. Deletion of RTS1, which encodes the only other known B type regulatory subunit of PP2A in S. cerevisiae, does not affect the bud morphology of the zds1Δ zds2Δ strains (Supplemental Figure 3). Thus, along with two-hybrid (Figure 4A) and macromolecular complex screening (Krogan et al., 2006; Collins et al., 2007) data, these data indicate that Cdc55p is the only PP2A regulatory B subunit that is physiologically relevant to Zds protein function.
Figure 5.
zds1Δ zds2Δ bud morphology defect requires CDC55. (A) Deletion of CDC55 in a zds1Δ zds2Δ strain suppressed the mutant, elongated bud phenotype at 30°C but not at 18°C. DIC micrographs of representative congenic, log-phase cdc55Δ (KKY1182), zds1Δ zds2Δ (KKY542), and cdc55Δ zds1Δ zds2Δ (KKY1200) cells cultured at 18 or 30°C in rich medium (YPD) for 4 h, after initial culturing at 30°C. Scale bar, 5 μm. (B) Quantification of mutant bud morphology in cultures of isogenic cdc55Δ zds1Δ zds2Δ strains containing (KKY1201; middle set of columns) or not containing (KKY1200; left set of columns) a CEN plasmid with CDC55. The KKY1201 strain was treated with 5-FOA to select for cells that had lost the CDC55 plasmid (right set of columns). Strains were grown as described in A. Elongated bud morphology was scored by DIC microscopy. n = 200 cells scored for each strain. Error bars, SD across three independent experiments.
Zds Proteins Negatively Regulate the Swe1-dependent G2/M Delay
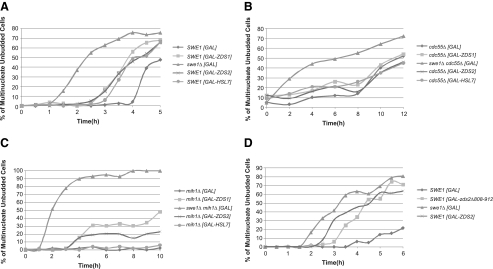
Although PP2A(Cdc55p) has been shown, among other processes, to act as a regulator of sister chromatid cohesion (Minshull et al., 1996) and mitotic exit (Queralt et al., 2006; Yellman and Burke, 2006; Queralt and Uhlmann, 2008), we reasoned that it was the less-characterized regulation of the Swe1p-dependent G2/M checkpoint (Wang and Burke, 1997; Yang et al., 2000; Pal et al., 2008) that was responsible for regulation of bud morphology by the Zds proteins. In other words, if the elongated bud morphology of the zds1Δ zds2Δ strain represents an overactive Swe1-dependent G2/M checkpoint, we hypothesized that the Zds proteins are negative regulators of the Swe1-dependent checkpoint, possibly through an interaction with Cdc55p. To test this hypothesis, we compared the rate of mitotic progression among cdc24-1ts cells expressing ZDS1, ZDS2, or HSL7 from a strong galactose-inducible promoter. The actin depolarization caused by cdc24-1ts at nonpermissive temperatures has been shown to delay mitotic progression in an SWE1-dependent manner (Adams and Pringle, 1984; Lew and Reed, 1995; McMillan et al., 1998; McMillan et al., 1999a). In this assay cultures were synchronized with α-factor at 25°C and then released from α-factor at 37°C, after which they were scored over time for the number of unbudded cells with two or more nuclei. This assay was used previously to define Hsl7p and Hsl1p as negative regulators of Swe1p (McMillan et al., 1999a). A SWE1 cdc24-1ts strain containing an empty GAL vector served as a baseline for the rate of mitotic progression when the Swe1p-dependent checkpoint is functional. As a control for mitotic progression in the absence of the checkpoint, we used a swe1Δ cdc24-1ts strain containing an empty GAL vector. In this assay, swe1Δ cells, which lack the checkpoint, are known to enter mitosis more rapidly than wild-type cells (McMillan et al., 1999a). We found that cdc24-1ts cells containing GAL-ZDS1 or GAL-ZDS2, grown in the presence of galactose, entered mitosis with the same kinetics as a cdc24-1ts strain containing GAL-HSL7 grown under the same conditions (Figure 6A). The rates of mitotic progression for these three strains were faster than that of a cdc24-1ts strain containing a GAL vector but slower than that of a swe1Δ cdc24-1ts strain containing the same vector. In a similar assay, ZDS1, ZDS2, and HSL7 also induced mitosis when the actin-depolymerizing drug latrunculin B was used to activate the checkpoint instead of cdc24-1ts (data not shown). These data indicate that the Zds proteins are negative regulators of the Swe1p-dependent G2/M checkpoint.
Figure 6.
ZDS genes negatively regulate the Swe1p-dependent G2/M checkpoint. Synchronized log-phase cultures of a congenic cdc24-1ts strains containing ZDS1, ZDS2, or HSL7 under the regulation of a strong galactose-inducible promoter were grown in rich medium (YP) containing 2% galactose and 2% raffinose and shifted to 37°C after release from α-factor arrest. Unbudded cells were scored for nuclear division. n = 200 cells were scored for each time point. Each experiment in A–D was replicated twice. (A) GAL-ZDS1 (KKY1184), GAL-ZDS2 (KKY1186), and GAL-HSL7 (KKY1187) promoted mitotic progression in cdc24-1ts cells at restrictive temperature. swe1Δ cdc24-1ts (KKY1185) and cdc24-1ts (KKY1183) cells contained an empty GAL vector and established baselines for mitotic progression without and with a functional checkpoint, respectively. (B) GAL-ZDS1 (KKY1205), GAL-ZDS2 (KKY1206), and GAL-HSL7 (KKY1207) failed to promote mitotic progression in cdc55Δ cdc24-1ts cells at restrictive temperature. cdc55Δ swe1Δ cdc24-1ts (KKY1203) and cdc55Δ cdc24-1ts (KKY1204) cells contained an empty GAL vector and established baselines for mitotic progression without and with a functional checkpoint, respectively. (C) GAL-ZDS1 (square; KKY1184), GAL-ZDS2 (KKY1186), but not GAL-HSL7 (KKY1187) promoted mitotic progression in mih1Δ cdc24-1ts cells at restrictive temperature. mih1Δ swe1Δ cdc24-1ts (KKY1192) and mih1Δ cdc24-1ts (KKY1188) cells contained an empty GAL vector and established baselines for mitotic progression without and with a functional checkpoint, respectively. (D) GAL-ZDS2 (KKY1186) and GAL-zds2Δ808-912 (KKY1217) promoted mitotic progression in cdc24-1ts cells. swe1Δ cdc24-1ts (KKY1185) and cdc24-1ts (KKY1183) cells contained an empty GAL vector and established baselines for mitotic progression without and with a functional checkpoint, respectively.
If Cdc55p activity is necessary for Zds-dependent regulation of the Swe1p-dependent checkpoint, the Zds proteins should be unable to promote mitosis in the absence of the CDC55 gene in the assay described above. We found that GAL-ZDS1, GAL-ZDS2, and GAL-HSL7 did not appreciably alter the kinetics of mitotic progression of a cdc55Δ cdc24-1ts strain compared with the same strain transformed with an empty GAL vector (Figure 6B). The results indicate that CDC55 is required for regulation of the Swe1p-dependent G2/M checkpoint in this assay.
Previous work by Pal et al. (2008) showed that deletion of CDC55 led to a change in the phosphorylation state of Mih1p. To determine whether the Zds proteins require Mih1p for regulation of the checkpoint, we repeated the assay in a cdc24-1ts strain lacking MIH1 (Figure 6C). We found ZDS1 and ZDS2 were able to induce mitosis, albeit at a rate slower than that observed previously for cells containing a functional MIH1 gene. In addition, to our surprise, we found that HSL7 required MIH1 to induce mitosis in this assay and a that the mih1Δ cdc24-1ts strain expressing an empty GAL vector cannot progress through mitosis. This observation is interesting because it implies that, once activated, the checkpoint stringently requires the Mih1p phosphatase for Hsl7p regulation of the checkpoint but the Zds proteins do not absolutely require Mih1p to have a regulatory effect.
If direct interaction of Zds2p with Cdc55p is necessary for regulation of the Swe1p-dependent checkpoint, we hypothesized that deletion of the ZH4 domain of Zds2p would impair the checkpoint regulation function of Zds2p. With the same mitotic progression assay used to show that Zds2p is a negative regulator of the Swe1p-dependent G2/M checkpoint, we found that strains containing GAL-ZDS2 (ZH4 domain present) and GAL-zds2Δ808-912 (ZH4 domain absent) exhibited similar kinetics of mitotic progression. This result indicated that Zds2p lacking the ZH4 domain is able to negatively regulate the Swe1p-dependent G2/M checkpoint (Figure 6D), meaning that Zds2p does not need to interact directly with Cdc55p to regulate the checkpoint. Because our mitotic progression assays (Figures 6,A–D) utilized a strain containing wild-type ZDS1 and ZDS2, we cannot exclude the alternate conclusion that the N-terminal region of Zds2p is a regulatory region, as suggested by Estruch et al. (2005), Schwer et al. (1998), and Yakura et al. (2006), and when under the control of the strong GAL promoter, it stimulates the regulatory activity of the endogenous Zds proteins present in the assay.
DISCUSSION
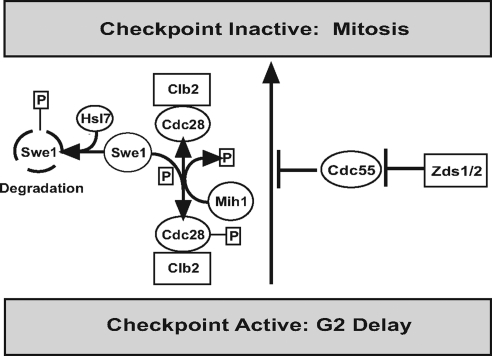
Herein, we showed that the paralogs ZDS1 and ZDS2 negatively regulate the Swe1p-dependent G2/M checkpoint and CDC55, which encodes a regulatory subunit of PP2A, is required for this regulation. Our structure–function mapping of Zds2p revealed a domain in the C-terminal region of Zds2p (ZH4) that is necessary for regulating polarized bud growth. These mapping data are consistent with a study by Schwer et al. (1998) that showed that either the C-terminus of Zds1p or Zds2p is sufficient for the rescue of the aberrant bud morphology of zds1Δ zds2Δ cells. In addition, by two-hybrid and pulldown assays, we showed that ZH4 is necessary and sufficient for the interaction of Zds2p with Cdc55p in vivo. Additionally we show, by in vitro-binding assay, that the C-terminus of Zds2p is sufficient for binding to recombinant Cdc55p. Rescue of the elongated bud morphology of zds1Δ zds2Δ cells by deletion of CDC55 demonstrated the physiological relevance of this interaction and suggests that Zds proteins are involved in a pathway that negatively regulates the Swe1p-dependent G2/M checkpoint in a Cdc55p-dependent manner (Figure 7) via the C-terminal ZH4 region of Zds2.
Figure 7.
Model of Zds protein regulation of the Swe1p-dependent G2/M checkpoint via Cdc55p. Cdc55p normally inhibits mitotic progression and the Zds proteins inhibit Cdc55p.
These results are important because the function of Zds1p and Zds2p has been enigmatic since their discovery (Bi and Pringle, 1996; Yu et al., 1996). Previous work revealed a broad spectrum of phenotypes when the ZDS genes are overexpressed or deleted (see Introduction). These studies did not reveal, however, a mechanism to satisfactorily explain the elongated bud phenotype of zds1Δ zds2Δ cells. Our study provides the first molecular link between the Zds proteins and the Swe1p-dependent G2/M checkpoint consistent with the zds1Δ zds2Δ phenotype.
Comparison of Zds2p with Zds1p
Because the ZH4 domains of Zds1p and Zds2p show 79% sequence identity (Supplemental Figure 1), we expect Cdc55p to bind Zds1p as well as Zds2p. Two lines of evidence support this idea. First, in high-throughput affinity interaction screens, both Zds1p and Zds2p interacted with subunits of the PP2A complex (Uetz et al., 2000; Drees et al., 2001; Gavin et al., 2002; Ho et al., 2002; Krogan et al., 2006; Collins et al., 2007). Second, in an accompanying study by John et al. (2011) it was shown by mass spectrometry that Zds1p bound to Cdc55p. It has not yet been shown formally, however, that the ZH4 domain of Zds1p binds Cdc55p directly.
As expected of functionally redundant proteins sharing a common binding partner, both Zds1p and Zds2p share a similar localization pattern. The cortical localization of Zds2p at the bud tip, shown in this study, is very similar to that of Zds1p (Bi and Pringle, 1996). Interestingly, Bi and Pringle (1996) also showed localization of Zds1p at the bud neck. We did not observe the same for Zds2p. One explanation for this discrepancy, rather than an actual difference in localization patterns between these proteins, is ectopic protein levels. We expressed epitope-tagged Zds2p with its endogenous promoter, whereas Bi and Pringle (1996) expressed GST-Zds1p with a strong promoter. They noted, however, that when GST-Zds1p was expressed at more physiological levels the localization at the bud neck disappeared. These localization data also differed from that of a proteome tagging study by Huh et al. (2003) in which both Zds1p and Zds2p localized diffusely throughout the cytoplasm. In our study, we did observe Zds2p in noncortical locations, but as puncta. When considering the formal possibility that those differences among the datasets is due to the type and position of tag used, it is important to note that our internally tagged Zds2p construct rescued the mutant morphology of zds1Δ zds2Δ cells (see Figure 3B). Considering that core regulatory elements of the Swe1p-dependent G2/M checkpoint exist in both nuclear and cytoplasmic space (Keaton et al., 2008), one possible function of the Zds proteins at the bud tip may be to provide sensors or scaffolds that link events at the site of polarized growth (e.g., organization of the actin cytoskeleton) to the nuclear mitotic cycle. We observed that a correlation between the localization of Zds2p at the apical bud tip and the ability to progress through mitosis is consistent with this idea.
Zds Proteins as Negative Regulators of the Swe1p-dependent G2/M Checkpoint
Although this study has identified a molecular function of the Zds proteins that explains the elongated bud phenotype in zds1Δ zds2Δ cells and expands our understanding of the regulatory inputs of the Swe1p-dependent G2/M checkpoint, the exact mechanism by which Zds proteins regulate the checkpoint in a Cdc55p-dependent manner remains unclear. Part of the difficulty in pinpointing the site of action of the Zds proteins is data that suggest Cdc55p and Zds1/2p possess additional functions in mitosis other than Swe1p-dependent G2/M (Wang and Burke, 1997; Kornitzer et al., 2001; Queralt et al., 2006; Yellman and Burke, 2006). For example, a study by Queralt et al. (2008) found that Zds1/2p may function in early anaphase to activate Cdc14p release from the nucleolus and thus activate the mitotic exit network. Alternatively, these observations might be explained by a Zds1/2p function in G2/M because both processes depend on the regulation of Cdc28p. Action of Cdc55p in anaphase does not easily explain our data showing a role for CDC55 in mitotic progression after checkpoint activation as Chiroli et al. (2007) elegantly showed that activation of the anaphase promoting complex is not sufficient for nuclear division in cells arrested by Swe1-dependent G2/M checkpoint activation. Our data do not preclude, however, multiple functions for a Zds/Cdc55p complex. If the Zds proteins are negative regulators of Cdc55p, as proposed by Queralt et al. (2008) and supported by our data, then overexpression of the Zds proteins might negatively regulate PP2A function in processes unrelated to the Swe1p-dependent G2/M checkpoint. Based on the ability of an SWE1 deletion to rescue the elongated bud morphology of a zds1Δ zds2Δ strain, it remains likely that the ability of Zds proteins to regulate Swe1p-dependent G2/M is physiologically relevant and does not represent a gain-of-function within our assays.
One mechanism by which the Zds proteins might be regulating the Swe1p checkpoint in a Cdc55p-dependent manner is through the Mih1p phosphatase, which opposes the Swe1p phosphorylation of Cdc28p. It has been proposed that the dephosphorylated form of Mih1p is the more active form (Pal et al., 2008) and that PP2A(Cdc55p) serves to activate Mih1p and thus negatively regulate the Swe1p-dependent G2/M checkpoint. Although the phosphorylation state of Mih1p is relevant toward understanding the Swe1p-dependent G2/M checkpoint in regards to its regulation by Zds proteins, phospho-mapping of Mih1p lies outside the scope of the current study. We do show, however, that the Zds proteins require MIH1 for efficient progress into mitosis when the checkpoint is activated. Considering that loss of Mih1p has no obvious effect on log-phase cells (Russell et al., 1989) even though it opposes the inhibitory phosphorylation of Cdc28p, we found it interesting that deletion of MIH1 completely blocked progression through the checkpoint (cf. Figure 6, A and C) when activated by cdc24-1ts. Because a small percentage of cells transformed with GAL-ZDS1 or GAL-ZDS2 escaped the checkpoint activation in an mih1Δ strain background (Figure 6C), it is possible that the Zds proteins possess an MIH1-independent function related to checkpoint inactivation, though we cannot rule out that the observed leakiness of the checkpoint is due to a gain-of-function upon strong ZDS1/2 expression. A MIH1-independent function is consistent with the idea that an alternate mechanism for dephosphorylation of Cdc28p exits (Pal et al., 2008). Another possibility is that the ZDS proteins regulate the checkpoint at the transcriptional or translational level. Sia et al. (1996) showed that inhibition of SWE1 transcription partially down-regulates the checkpoint. Additionally, Ma et al. (1996) showed that deletion of ZDS1 (OSS1) causes the SWE1 transcript level, which normally exhibits cell cycle–dependent transcription, to partially decouple from the cell cycle and remain high during G2 and M phases. Ma et al. (1996) concluded that ZDS1 is necessary for repression of SWE1 transcripts, which might explain why the ZDS genes, when induced, can down-regulate the checkpoint in an MIH1 deletion strain but that HSL7, which works at the protein level to degrade Swe1p, cannot. Looking at the larger picture, our data do not preclude the Zds proteins from functioning both pre- or posttranslationally, as suggested by Ma et al. (1996) and Pal et al. (2008), respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Longtine (Washington University, St. Louis, MO), B. Stillman (Cold Spring Harbor Laboratories, Cold Spring Harbor, NY), and E. Bi (University of Pennsylvania, Philadelphia, PA) for the generous gift of plasmids and D. Lew (Duke University, Durham, NC) and D. Drubin (University of California, Berkeley, CA) for the generous gift of strains. We especially thank Doug Kellogg (University of California, Santa Cruz, CA) and his lab for sharing unpublished results and for critical comments on the manuscript. Parts of this work were completed by K.M.Y. in partial fulfillment of the requirements for the degree Doctor of Philosophy (University of Virginia) and was supported by National Institute of General Medical Sciences Training Grant (T32 GM08136) award to K.M.Y., National Science Foundation Grant 0723342 to K.G.K., and National Institutes of Health Grant P41 RR11823 to S.F.
Abbreviation used:
- ZH4
Zds homology 4.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-04-0326) on October 27, 2010.
REFERENCES
- Adams A. E., Pringle J. R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amon A., Surana U., Muroff I., Nasmyth K. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature. 1992;355:368–371. doi: 10.1038/355368a0. [DOI] [PubMed] [Google Scholar]
- Bandhakavi S., McCann R. O., Hanna D. E., Glover C. V. Genetic interactions among ZDS1,2, CDC37, and protein kinase CK2 in Saccharomyces cerevisiae. FEBS Lett. 2003;554:295–300. doi: 10.1016/s0014-5793(03)01165-7. [DOI] [PubMed] [Google Scholar]
- Bi E., Pringle J. R. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialojan C., Takai A. Inhibitory effect of a marine-sponge toxin, okadaic acid, on protein phosphatases. Specificity and kinetics. Biochem. J. 1988;256:283–290. doi: 10.1042/bj2560283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher R. N., Deshaies R. J., Kirschner M. W. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher N., Britten C. D. G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. Br. J. Cancer. 2008;98:523–528. doi: 10.1038/sj.bjc.6604208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiroli E., Rossio V., Lucchini G., Piatti S. The budding yeast PP2ACdc55 protein phosphatase prevents the onset of anaphase in response to morphogenetic defects. J. Cell Biol. 2007;177:599–611. doi: 10.1083/jcb.200609088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. R., Kemmeren P., Zhao X. C., Greenblatt J. F., Spencer F., Holstege F. C., Weissman J. S., Krogan N. J. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell Proteom. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Dasso M., Newport J. W. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: studies in Xenopus. Cell. 1990;61:811–823. doi: 10.1016/0092-8674(90)90191-g. [DOI] [PubMed] [Google Scholar]
- Dighe S. A., Kozminski K. G. Swf1p, a member of the DHHC-CRD family of palmitoyltransferases, regulates the actin cytoskeleton and polarized secretion independently of its DHHC motif. Mol. Biol. Cell. 2008;19:4454–4468. doi: 10.1091/mbc.E08-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees B. L., et al. A protein interaction map for cell polarity development. J. Cell Biol. 2001;154:549–571. doi: 10.1083/jcb.200104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy W. G., Kumagai A. The cdc25 protein contains an intrinsic phosphatase activity. Cell. 1991;67:189–196. doi: 10.1016/0092-8674(91)90582-j. [DOI] [PubMed] [Google Scholar]
- Eichhorn P. J., Creyghton M. P., Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim. Biophys. Acta. 2009;1795:1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Enoch T., Nurse P. Mutation of fission yeast cell cycle control genes abolishes dependence of mitosis on DNA replication. Cell. 1990;60:665–673. doi: 10.1016/0092-8674(90)90669-6. [DOI] [PubMed] [Google Scholar]
- Enserink J. M., Smolka M. B., Zhou H., Kolodner R. D. Checkpoint proteins control morphogenetic events during DNA replication stress in Saccharomyces cerevisiae. J. Cell Biol. 2006;175:729–741. doi: 10.1083/jcb.200605080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F., Hodge C. A., Rodriguez-Navarro S., Cole C. N. Physical and genetic interactions link the yeast protein Zds1p with mRNA nuclear export. J. Biol. Chem. 2005;280:9691–9697. doi: 10.1074/jbc.M413025200. [DOI] [PubMed] [Google Scholar]
- Gauss R., Trautwein M., Sommer T., Spang A. New modules for the repeated internal and N-terminal epitope tagging of genes in Saccharomyces cerevisiae. Yeast. 2005;22:1–12. doi: 10.1002/yea.1187. [DOI] [PubMed] [Google Scholar]
- Gautier J., Solomon M. J., Booher R. N., Bazan J. F., Kirschner M. W. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 1991;67:197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- Gavin A. C., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gould K. L., Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- Griffioen G., Branduardi P., Ballarini A., Anghileri P., Norbeck J., Baroni M. D., Ruis H. Nucleocytoplasmic distribution of budding yeast protein kinase A regulatory subunit Bcy1 requires Zds1 and is regulated by Yak1-dependent phosphorylation of its targeting domain. Mol. Cell. Biol. 2001;21:511–523. doi: 10.1128/MCB.21.2.511-523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Culotti J., Pringle J. R., Reid B. J. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Healy A. M., Zolnierowicz S., Stapleton A. E., Goebl M., DePaoli-Roach A. A., Pringle J. R. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol. Cell. Biol. 1991;11:5767–5780. doi: 10.1128/mcb.11.11.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo S. J., Tatebayashi K., Ikeda H. The budding yeast cohesin gene SCC1/MCD1/RHC21 genetically interacts with PKA, CDK and APC. Curr. Genet. 1999;36:329–338. doi: 10.1007/s002940050507. [DOI] [PubMed] [Google Scholar]
- Ho Y., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hook S. S., Lin J. J., Dutta A. Mechanisms to control rereplication and implications for cancer. Curr. Opin. Cell Biol. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. L., Chen Y. S., Tsai S. Y., Tu P. J., Wang M. J., Lin J. J. Interaction of Saccharomyces Cdc13p with Pol1p, Imp4p, Sir4p and Zds2p is involved in telomere replication, telomere maintenance and cell growth control. Nucleic Acids Res. 2004;32:511–521. doi: 10.1093/nar/gkh203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V., Longin S., Goris J. PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail) Trends Biochem. Sci. 2008;33:113–121. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- John S. W., Tjandra H., Schieltz D., Yates J., III, Kellogg D. R. The Zds proteins control entry into mitosis and target protein phosphatase 2A to the Cdc25 phosphatase. Mol. Biol. Cell. 2011 doi: 10.1091/mbc.E10-06-0487. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaton M. A., Szkotnicki L., Marquitz A. R., Harrison J., Zyla T. R., Lew D. J. Nucleocytoplasmic trafficking of G2/M regulators in yeast. Mol. Biol. Cell. 2008;19:4006–4018. doi: 10.1091/mbc.E08-03-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornitzer D., Sharf R., Kleinberger T. Adenovirus E4orf4 protein induces PP2A-dependent growth arrest in Saccharomyces cerevisiae and interacts with the anaphase-promoting complex/cyclosome. J. Cell Biol. 2001;154:331–344. doi: 10.1083/jcb.200104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K. G., Alfaro G., Dighe S., Beh C. T. Homologues of oxysterol-binding proteins affect Cdc42p- and Rho1p-mediated cell polarization in Saccharomyces cerevisiae. Traffic. 2006;7:1224–1242. doi: 10.1111/j.1600-0854.2006.00467.x. [DOI] [PubMed] [Google Scholar]
- Kozminski K. G., Chen A. J., Rodal A. A., Drubin D. G. Functions and functional domains of the GTPase Cdc42p. Mol. Biol. Cell. 2000;11:339–354. doi: 10.1091/mbc.11.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N. J., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W. G. Regulation of the cdc25 protein during the cell cycle in Xenopus extracts. Cell. 1992;70:139–151. doi: 10.1016/0092-8674(92)90540-s. [DOI] [PubMed] [Google Scholar]
- Larkin M. A., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J. Cell Biol. 1995;129:739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobrich M., Jeggo P. A. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lundgren K., Walworth N., Booher R., Dembski M., Kirschner M., Beach D. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell. 1991;64:1111–1122. doi: 10.1016/0092-8674(91)90266-2. [DOI] [PubMed] [Google Scholar]
- Lydall D., Nikolsky Y., Bishop D. K., Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- Ma X. J., Lu Q., Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- Majka J., Burgers P. M. Yeast Rad17/Mec3/Ddc1, a sliding clamp for the DNA damage checkpoint. Proc. Natl. Acad. Sci. USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleverty C. J., Hornsby M., Spraggon G., Kreusch A. Crystal structure of human Pus10, a novel pseudouridine synthase. J. Mol. Biol. 2007;373:1243–1254. doi: 10.1016/j.jmb.2007.08.053. [DOI] [PubMed] [Google Scholar]
- McMillan J. N., Longtine M. S., Sia R. A., Theesfeld C. L., Bardes E. S., Pringle J. R., Lew D. J. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol. Cell. Biol. 1999a;19:6929–6939. doi: 10.1128/mcb.19.10.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J. N., Sia R. A., Bardes E. S., Lew D. J. Phosphorylation-independent inhibition of Cdc28p by the tyrosine kinase Swe1p in the morphogenesis checkpoint. Mol. Cell. Biol. 1999b;19:5981–5990. doi: 10.1128/mcb.19.9.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J. N., Sia R. A., Lew D. J. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J. Cell Biol. 1998;142:1487–1499. doi: 10.1083/jcb.142.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty J. J., Lew D. J. Swe1p responds to cytoskeletal perturbation, not bud size, in S. cerevisiae. Curr. Biol. 2005;15:2190–2198. doi: 10.1016/j.cub.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Minshull J., Straight A., Rudner A. D., Dernburg A. F., Belmont A., Murray A. W. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 1996;6:1609–1620. doi: 10.1016/s0960-9822(02)70784-7. [DOI] [PubMed] [Google Scholar]
- Miranda T. B., Sayegh J., Frankel A., Katz J. E., Miranda M., Clarke S. Yeast Hsl7 (histone synthetic lethal 7) catalyses the in vitro formation of omega-N(G)-monomethylarginine in calf thymus histone H2A. Biochem. J. 2006;395:563–570. doi: 10.1042/BJ20051771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Nurse P., Russell P. Regulation of mitosis by cyclic accumulation of p80cdc25 mitotic inducer in fission yeast. Nature. 1990;344:549–552. doi: 10.1038/344549a0. [DOI] [PubMed] [Google Scholar]
- Notredame C., Higgins D. G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- Ohnuki S., Nogami S., Kanai H., Hirata D., Nakatani Y., Morishita S., Ohya Y. Diversity of Ca2+-induced morphology revealed by morphological phenotyping of Ca2+-sensitive mutants of Saccharomyces cerevisiae. Eukaryot. Cell. 2007;6:817–830. doi: 10.1128/EC.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal G., Paraz M. T., Kellogg D. R. Regulation of Mih1/Cdc25 by protein phosphatase 2A and casein kinase 1. J. Cell Biol. 2008;180:931–945. doi: 10.1083/jcb.200711014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierstorff E., Kane C. M. Genetic interactions between an RNA polymerase II phosphatase and centromeric elements in Saccharomyces cerevisiae. Mol. Genet. Genom. 2004;271:603–615. doi: 10.1007/s00438-004-1009-5. [DOI] [PubMed] [Google Scholar]
- Queralt E., Lehane C., Novak B., Uhlmann F. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell. 2006;125:719–732. doi: 10.1016/j.cell.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Queralt E., Uhlmann F. Separase cooperates with Zds1 and Zds2 to activate Cdc14 phosphatase in early anaphase. J. Cell Biol. 2008;182:873–883. doi: 10.1083/jcb.200801054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N., Runge K. W. The ZDS1 and ZDS2 proteins require the Sir3p component of yeast silent chromatin to enhance the stability of short linear centromeric plasmids. Chromosoma. 1999;108:146–161. doi: 10.1007/s004120050364. [DOI] [PubMed] [Google Scholar]
- Roy N., Runge K. W. Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr. Biol. 2000;10:111–114. doi: 10.1016/s0960-9822(00)00298-0. [DOI] [PubMed] [Google Scholar]
- Russell P., Moreno S., Reed S. I. Conservation of mitotic controls in fission and budding yeasts. Cell. 1989;57:295–303. doi: 10.1016/0092-8674(89)90967-7. [DOI] [PubMed] [Google Scholar]
- Sayegh J., Clarke S. G. Hsl7 is a substrate-specific type II protein arginine methyltransferase in yeast. Biochem. Biophys. Res. Commun. 2008;372:811–815. doi: 10.1016/j.bbrc.2008.05.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Linder P., Shuman S. Effects of deletion mutations in the yeast Ces1 protein on cell growth and morphology and on high copy suppression of mutations in mRNA capping enzyme and translation initiation factor 4A. Nucleic Acids Res. 1998;26:803–809. doi: 10.1093/nar/26.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya-Kawasaki M., Abe M., Saka A., Watanabe D., Kono K., Minemura-Asakawa M., Ishihara S., Watanabe T., Ohya Y. Dissection of upstream regulatory components of the Rho1p effector, 1,3-beta-glucan synthase, in Saccharomyces cerevisiae. Genetics. 2002;162:663–676. doi: 10.1093/genetics/162.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. B. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Shinohara M., Sakai K., Ogawa T., Shinohara A. The mitotic DNA damage checkpoint proteins Rad17 and Rad24 are required for repair of double-strand breaks during meiosis in yeast. Genetics. 2003;164:855–865. doi: 10.1093/genetics/164.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., Yang H., Hallberg E., Hallberg R. Molecular genetic analysis of Rts1p, a B' regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol. Cell. Biol. 1997;17:3242–3253. doi: 10.1128/mcb.17.6.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia R. A., Bardes E. S., Lew D. J. Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J. 1998;17:6678–6688. doi: 10.1093/emboj/17.22.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia R. A., Herald H. A., Lew D. J. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol. Biol. Cell. 1996;7:1657–1666. doi: 10.1091/mbc.7.11.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P. K., Murray A. W. S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc28. Nature. 1992;355:365–368. doi: 10.1038/355365a0. [DOI] [PubMed] [Google Scholar]
- Suganuma M., Fujiki H., Suguri H., Yoshizawa S., Hirota M., Nakayasu M., Ojika M., Wakamatsu K., Yamada K., Sugimura T. Okadaic acid: an additional non-phorbol-12-tetradecanoate-13-acetate-type tumor promoter. Proc. Natl. Acad. Sci. USA. 1988;85:1768–1771. doi: 10.1073/pnas.85.6.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Nojima H. Nik1, a Nim1-like protein kinase of S. cerevisiae interacts with the Cdc28 complex and regulates cell cycle progression. Genes Cells. 1996;1:905–921. doi: 10.1046/j.1365-2443.1996.d01-213.x. [DOI] [PubMed] [Google Scholar]
- Tong A. H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Uetz P., et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wang Y., Burke D. J. Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:620–626. doi: 10.1128/mcb.17.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakura M., Ozoe F., Ishida H., Nakagawa T., Tanaka K., Matsuda H., Kawamukai M. zds1, a novel gene encoding an ortholog of Zds1 and Zds2, controls sexual differentiation, cell wall integrity and cell morphology in fission yeast. Genetics. 2006;172:811–825. doi: 10.1534/genetics.105.050906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Jiang W., Gentry M., Hallberg R. L. Loss of a protein phosphatase 2A regulatory subunit (Cdc55p) elicits improper regulation of Swe1p degradation. Mol. Cell. Biol. 2000;20:8143–8156. doi: 10.1128/mcb.20.21.8143-8156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellman C. M., Burke D. J. The role of Cdc55 in the spindle checkpoint is through regulation of mitotic exit in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006;17:658–666. doi: 10.1091/mbc.E05-04-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H., Mizunuma M., Okamoto M., Yamamoto J., Hirata D., Miyakawa T. Involvement of calcineurin-dependent degradation of Yap1p in Ca2+-induced G2 cell-cycle regulation in Saccharomyces cerevisiae. EMBO Rep. 2006;7:519–524. doi: 10.1038/sj.embor.7400647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Jiang Y. W., Wellinger R. J., Carlson K., Roberts J. M., Stillman D. J. Mutations in the homologous ZDS1 and ZDS2 genes affect cell cycle progression. Mol. Cell. Biol. 1996;16:5254–5263. doi: 10.1128/mcb.16.10.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli C. F., Valentini S. R. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics. 2005;171:1571–1581. doi: 10.1534/genetics.105.048082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.