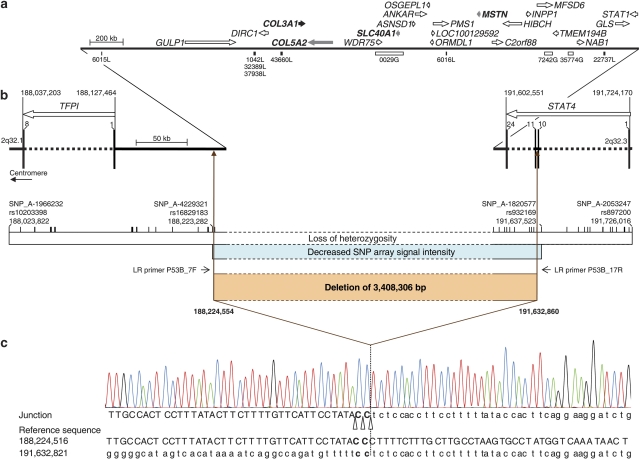
Figure 1.
Deletion of 3.4 Mb identified in this study. (a) Schematic representation of the completely deleted genes. Genes are represented by arrows that indicate the direction of transcription according to Entrez Gene (www.ncbi.nlm.nih.gov/sites/entrez?db=gene; version May 2009). COL3A1 is denoted by a black arrow and other genes associated with a dominantly inherited disease and thus considered for further analyses are indicated by gray arrows. Known copy number variations (CNVs) in this region are given below the line according to the Database of Genomic Variants (DGV, http://projects.tcag.ca/variation, version May 2009; Supplementary Table S3). The black bars represent losses (L) and white bars gains (G). (b) Schematic representation of the region flanking the breakpoints. Exons are specified as bars and marked with the corresponding number. Regions derived from high-density microarray analyses are represented by a white bar for loss of heterozygosity and by a blue bar for decreased SNP array signal intensity. The positions of SNPs tested by the array set are indicated by vertical lines. The deleted region is denoted as a brown bar and the primers used for long-range (LR) PCR (LR primers P53B_7F and P53B_17R) are indicated by arrows. (c) Sequence of the long-range PCR product spanning the breakpoint junction of the deletion. Uppercase letters represent the sequence in the region of the start point of the deletion and lowercase letters the sequence in the region of the deletion end point. Because of identical sequences at the site of the breakpoints, the break and rejoining could have occurred at three positions as indicated by open triangles. The dotted line marks the most telomeric position of the possible breakpoints. All nucleotide positions are given in relation to the human genome reference sequence (NCBI build 36.1, March 2006).