Figure 2.
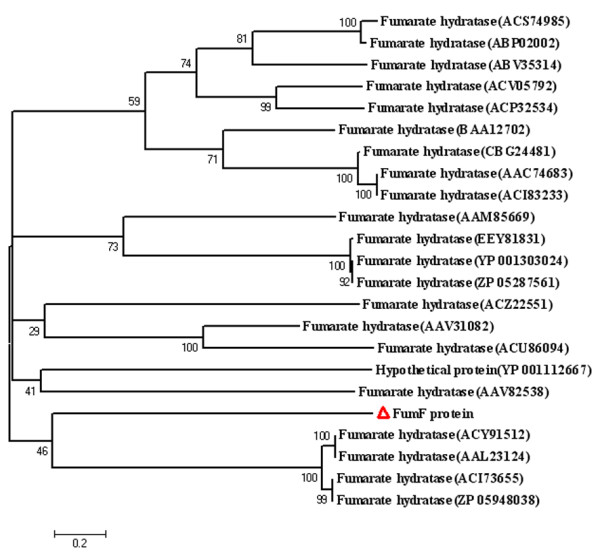
Phylogenetic relationship of FumF protein with other fumarases. Sequence alignment was performed using ClustalW version 1.81, and the phylogenetic tree was constructed by the neighbor-joining method using MEGA version 4.0 [30]. Boot-strapping values were used to estimate the reliability of phylogenetic reconstructions (1,000 replicates). The numbers associated with the branches refer to bootstrap values (confidence limits) representing the substitution frequencies per amino acid residue. Fumarases are identified by their GenBank accession numbers.
