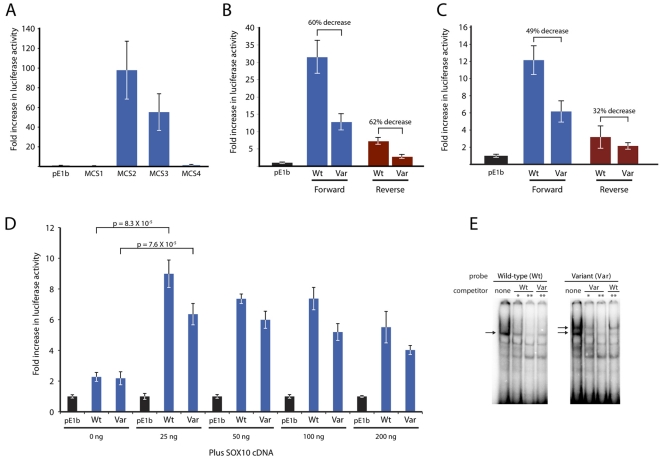
Figure 2. Functional characterization of the c.126-1086T>A variant in MPZ-MCS3.
(A) Genomic segments spanning each MPZ MCS (MPZ-MCS1, MPZ-MCS2, MPZ-MCS3, and MPZ-MCS4) were tested for their ability to direct luciferase reporter-gene expression in cultured Schwann (S16) cells compared to a control vector (pE1b). Error bars indicate standard deviation. (B) Wild-type (Wt) and c.126-1086T>A variant (Var) forms of MPZ-MCS3 were tested for their ability to direct luciferase reporter-gene expression in the forward (blue) and reverse (red) orientations in cultured Schwann (S16) cells compared to a control vector (pE1b). Error bars indicate standard deviation. (C) Similar experiments as described in B were performed with cultured oligodendrocytes (OliNeu) cells. (D) Wild-type (Wt) and c.126-1086T>A (Var) forms of MPZ-MCS3 were tested for their responsiveness to SOX10 via luciferase reporter-gene expression in SOX10-negative MN1 cells, in the forward orientation compared to a control vector (pE1b). The amount of SOX10 expression vector co-transfected is shown. Error bars indicate standard deviation and p-values were calculated using the student's t-test. (E) Electrophoretic mobility shift analysis (EMSA) of wild-type and c.126-1086T>A alleles. ‘Probes’ (radiolabeled oligonucleotides) and ‘competitors’ (unlabeled oligonucleotides) corresponding to a 25-bp fragment including the wild-type (Wt) and c.126-1086T>A variant-containing (Var) consensus SOX10-binding sequence in MPZ-MCS3 (see Fig. 1B) were incubated with S16 nuclear extracts and subjected to electrophoresis (see Materials and Methods). Each assay used the same amount of probe coupled with varying amounts of competitor, as shown (‘none’ indicates no competitor; * and ** indicate a probe to competitor ratio of 1∶10 and 1∶100, respectively). Arrows denote specific nuclear proteins bound to the probe.