Figure 5. ESC genes show distinct histone acetylation patterns.
We compare five sets of ESC specific genes to all other genes in terms of their histone acetylation and gene expression changes: (1) members of the PluriNet [45]; (2) hits of a recent RNAi screen [46]; (3) gene ontology term GO:0019827 ‘stem cell maintenance’; (4) members of an ESC-specific protein-interaction network [15]; (5) key transcriptional regulators [8]. A All genes are ordered by their mean acetylation signal on day 0, their acetylation change on day 5 and their expression change on day 5. The positions of the five ES specific gene sets in this ordering are then indicated by bars. The dots and circles indicate statistical significance of observed trends evaluted by GSEA: three dots for  , two for
, two for  and one for
and one for  , while a circle represents
, while a circle represents 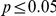 . B Here we compare ES genes to all others over the whole acetylation profile. The blue areas indicate quantiles of the genome-wide distribution of acetylation signal. The ES specific gene sets (white boxplots) show overall very high acetylation levels, in particular the transcriptional regulators (red dots) show surprisingly high histone acetylation levels before TSS.
. B Here we compare ES genes to all others over the whole acetylation profile. The blue areas indicate quantiles of the genome-wide distribution of acetylation signal. The ES specific gene sets (white boxplots) show overall very high acetylation levels, in particular the transcriptional regulators (red dots) show surprisingly high histone acetylation levels before TSS.

