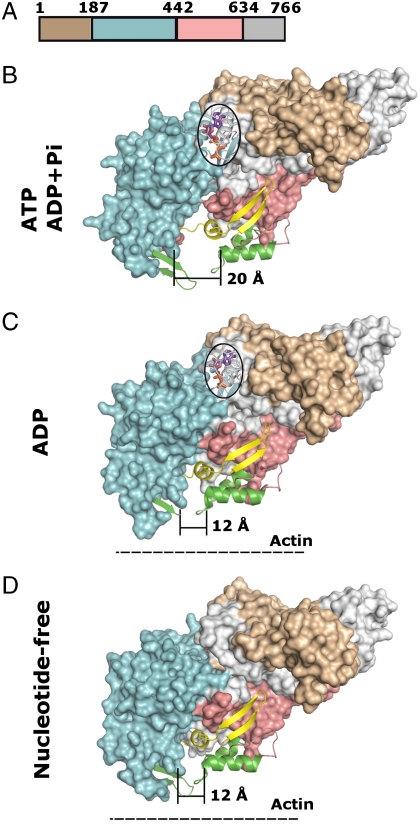
Fig. 5.
Projection of She4p binding sites onto the myosin surface during the ATPase cycle. (A) Location of modules in the primary structure of chicken myosin V motor domain; residue numbers are indicated and domain boundaries are colored: brown, N-terminal region; cyan, Upper subdomain; pink, Lower subdomain; and gray, C-terminal region. (B–D) Surface representation of the motor domain of chicken myosin V in the ATP or ADP + Pi form [Protein Data Bank (PDB) ID code: 1w7j(B)], in the ADP form [PDB ID code: 1w7i(c)], or in the nucleotide-free form [PDB ID code: 1w8j(D)], respectively, (27); color code as in (A). Note the dramatic reduction from 20 to 12 Å in width at the base of the cleft during the ATPase cycle, concomitant with strong binding to actin filaments (dashed line). Projected on the surface of the myosin forms are nucleotides (magenta, stick and ball representation), strong actin binding sites (green, ribbon representation), and the UCS binding sites (yellow, ribbon representation). Note that the UCS binding site contains two distinct binding epitopes; One, consisting of two antiparallel beta strands, remains always exposed on the myosin surface during the ATPase cycle, while the other one, located inside the cleft (helix loop) is occluded during the reduction of the width of the cleft from 20 to 12 Å during the ATPase cycle.