Abstract
Following its tyrosine phosphorylation, STAT3 is methylated on K140 by the histone methyl transferase SET9 and demethylated by LSD1 when it is bound to a subset of the promoters that it activates. Methylation of K140 is a negative regulatory event, because its blockade greatly increases the steady-state amount of activated STAT3 and the expression of many (i.e., SOCS3) but not all (i.e., CD14) STAT3 target genes. Biological relevance is shown by the observation that overexpression of SOCS3 when K140 cannot be methylated blocks the ability of cells to activate STAT3 in response to IL-6. K140 methylation does not occur with mutants of STAT3 that do not enter nuclei or bind to DNA. Following treatment with IL-6, events at the SOCS3 promoter occur in an ordered sequence, as shown by chromatin immunoprecipitations. Y705-phosphoryl-STAT3 binds first and S727 is then phosphorylated, followed by the coincident binding of SET9 and dimethylation of K140, and lastly by the binding of LSD1. We conclude that the lysine methylation of promoter-bound STAT3 leads to biologically important down-regulation of the dependent responses and that SET9, which is known to help provide an activating methylation mark to H3K4, is recruited to the newly activated SOCS3 promoter by STAT3.
Keywords: gene regulation, signal transduction, gene regulation, posttranslational modifications
Covalent modifications of histones and nonhistone chromatin proteins are essential for control of gene expression. The N-terminal tails of histones, as well as positions in the globular domains, carry posttranslational modifications, such as methylation, acetylation, phosphorylation, ADP ribosylation, and ubiquitination (1). Multiple residues of each of the four core histones have been identified as modification sites (2) and some of the same lysine side chains can be either methylated or acetylated. These modifications alter chromatin structure, often by providing entry sites for proteins that determine higher-order chromatin organization, leading to the activation or inactivation of specific genes. In addition, methylation and demethylation of p53 and NFκB are carried out by enzymes previously known to modify only histones. For p53, the reactions occur on K370, K372, and K382 (3). For NFκB, K37 is methylated by SET9 (4), and K218 and K221 are methylated by NSD1 and demethylated by FBXL11 (5).
STAT3 is phosphorylated on tyrosine and serine residues in response to many different cytokines and growth factors, leading to the formation of dimers through reciprocal phosphotyrosine–SH2 interactions (6). Activated STAT3 dimers bind to and activate the promoters of target genes. In addition to phosphorylation, STAT3 was reported to be acetylated at K685 following cytokine stimulation, and the K685R mutation blocked its activation (7), but these observations have been disputed (8). Ray et al. (9) reported that K49 and K87 of STAT3 are acetylated by p300 and that the K-R mutations resulted in a STAT3 protein that is able to translocate into nuclei, but unable to bind to p300. Here, we show that, in response to IL-6, STAT3 is methylated on K140 by the H3K4 methyl transferase SET9 and demethylated by the H3K4 demethylase LSD1 (lysine-specific demethylase 1, also named BHC110). Prevention of methylation by mutation of K140 greatly enhances the induction of one group of genes in response to IL-6, but has little effect on a second group, and inhibits the activation of a third group. Several lines of evidence indicate that methylation takes place as STAT3 is bound to promoters in the first but not the second group of genes.
Results
MS Indicates that STAT3 Is Dimethylated on K140.
To purify STAT3 for analysis, we generated the STAT3-null human cell line A4 by homologous recombination and expressed Flag-tagged STAT3 at a level comparable to that in parental DLD1 cells. In A4-STAT3 cells, the tagged STAT3 responded to IL-6 normally (Fig. S1A). STAT3 was purified from untreated or IL-6–treated A4-STAT3 cells (Fig. S1A). The samples were analyzed by MS. Mass shifts of +14 and +28 were observed for a STAT3 peptide from treated cells only (Fig. S1 B–D), signifying both mono- and dimethyl modifications. However, the amount of monomethyl STAT3 was far less than that of dimethyl STAT3. The methylations were in a peptide that includes residues 115 to 140, and tandem MS analysis localized the site to residues 131 to 140. Because K140 is the most logical residue in this peptide for modification, we generated antibodies against STAT3 peptides with K140me1 and K140me2. The specificity of each antibody was confirmed by ELISA and Western analyses (Fig. S2 A and B). Using these antibodies, there was a strong signal for STAT3-K140me2 and no signal for STAT3-K140me1 in IL-6–treated parental DLD1 cells and A4-STAT3 cells (Fig. 1A, third panel).
Fig. 1.
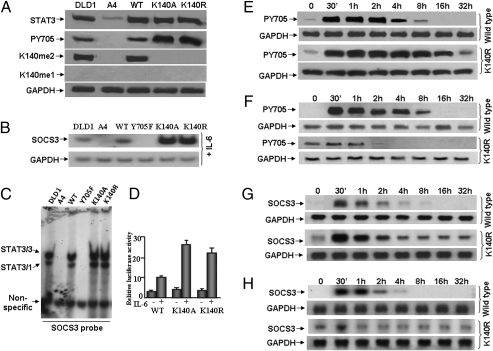
K140A or K140R mutation of STAT3 increases its tyrosine phosphorylation and transcriptional activity in response to IL-6. (A) Western analyses for STAT3 and Y705-phosphoryl-STAT3. STAT3-null A4 cells were infected with retroviral constructs and stable pools of cells were selected with G418. DLD1, parental colon cancer cells; A4, STAT3-null cells; WT, A4 cells expressing a normal level of wild-type STAT3; K140A, A4 cells expressing a normal level of K140A STAT3; K140R, A4 cells expressing a normal level of K140R STAT3. The cells were treated with IL-6 for 4 h and total cell lysates were analyzed by Western blot. (B) Northern analysis of gene expression. Cells were treated with IL-6 for 4 h and total RNA was analyzed for GAPDH and SOCS3 mRNAs. (C) EMSAs. Whole-cell extracts were made from DLD1 cells, A4 cells, or A4 cells expressing wild-type, Y705F, K140A, or K140R STAT3, treated with IL-6 for 4 h. A GAS (STAT3 binding) element derived from the SOCS3 promoter was used as the probe. (D) SOCS3 reporter gene induction. Luciferase constructs containing ∼1 kb of the human SOCS3 promoter (from −1 to about −1,000) were cotransfected with a pCH110 control plasmid. Cells stimulated with IL-6 for 4 h or untreated cells were washed with serum-free medium and cultured in complete medium for 12 h more. Cell lysates were analyzed for luciferase activity, which was normalized to the level of β-galactosidase activity from pCH110 control cells in the same extract. Values are means of triplicate determinations and the bars show one SEM. (E) A4 cells expressing wild-type or K140R STAT3 were treated with IL-6 for 4 h. The cells were washed with serum-free medium twice and cultured in complete medium without IL-6. Whole-cell lysates were analyzed by Western blot for total STAT3 and Y705-phosphoryl-STAT3. (F) A4 cells expressing wild-type or K140R STAT3 were treated with IL-6 for 4 h. The cells were washed with serum-free medium twice and cultured in complete medium without IL-6. After 32 h, the above cells were retreated with IL-6 for 4 h, and then washed with serum-free medium twice and cultured in complete medium, again without IL-6. Whole-cell lysates were analyzed by Western blot for total STAT3 and Y705-phosphoryl-STAT3. (G) A4 cells expressing wild-type or K140R-STAT3 were treated with IL-6 for 4 h. The cells were washed with serum-free medium twice and cultured in complete medium without IL-6. Total RNA was analyzed for GAPDH and SOCS3 mRNAs. (H) A4 cells expressing wild-type or K140R STAT3 were treated with IL-6 for 4 h. The cells were washed with serum-free medium twice and cultured in complete medium without IL-6. After 32 h, the cells were retreated with IL-6 for 4 h, washed with serum-free medium twice and cultured in complete medium without IL-6. Total RNA was analyzed for GAPDH and SOCS3 mRNAs.
Mutation of K140 of STAT3 Prolongs Its Transcriptional Activity in Response to IL-6.
STAT3 proteins with K140R or K140A mutations were expressed stably at normal levels in A4 cells (Fig. 1A, first panel). As expected, cells expressing the mutant proteins did not show methylation of K140 in response to IL-6 (Fig. 1A, third panel). In addition, cells containing the mutant proteins showed much more phosphorylation of Y705 in response to IL-6 than did cells expressing similar levels of wild-type STAT3 (Fig. 1A, second panel). To determine how gene expression might be affected by K140 mutations, we measured the effect on SOCS3, an important direct target of STAT3 that encodes a potent negative regulator of STAT3 activation. In response to IL-6, the level of SOCS3 mRNA increased dramatically in cells expressing the K140R or K140A mutants (Fig. 1B). EMSAs showed that both mutations enhanced the ability of STAT3 to bind to the GAS element of the SOCS3 promoter (Fig. 1C). SOCS3 promoter-driven luciferase activity was substantially higher in cells expressing the K140R or K140A mutant proteins than in cells expressing wild-type STAT3 (Fig. 1D). The enhanced binding of STAT3 to DNA (Fig. 1C) and luciferase reporter activation (Fig. 1D) are a result of enhanced activation of the K140R mutant protein. The mechanistic connection between lysine methylation and the degree of phosphorylation of tyrosine 705 is not yet clear.
An assay for the stability of Y705 phosphorylation was performed by treating cells with IL-6 for 4 h, followed by the removal of IL-6. The phosphorylation of Y705 of STAT3 was sustained for much longer in cells expressing the K140R (Fig. 1E) mutant protein than in cells expressing wild-type STAT3. To test the physiological significance of the dimethylation of K140, A4 cells expressing wild-type or K140R STAT3 were treated with IL-6 for 4 h, kept without IL-6 for 32 h, and then treated again with IL-6. The cells expressing wild-type STAT3 responded to the second IL-6 treatment normally, by phosphorylating Y705 (Fig. 1F, first panel). However, the cells expressing K140R STAT3 failed to respond to the second treatment with IL-6 (Fig. 1F, third panel). The SOCS3 gene responds to the first IL-6 treatment in cells with either wild-type or K140R STAT3 (Fig. 1G) but to the second IL-6 treatment only in cells with wild-type STAT3 (Fig. 1H). These results show that the dimethylation of STAT3 is required to maintain normal cellular responsiveness to cytokines, at least in part by reducing the duration and extent of expression of the potent negative-regulator SOCS3. In addition, the dimethylation of STAT3 is induced by a variety of stimuli (Fig. S3) (i.e., oncostatin M, EGF in human breast epithelial hTERT-HME1 cells, or by IFN in cells that lack STAT1) (10). Therefore, activation of STAT3 by several different means induces the formation of STAT3-K140me2 similarly.
Nuclear Import and DNA Binding Are Required for IL-6–induced S727 Phosphorylation and K140 Dimethylation of STAT3.
To determine whether nuclear translocation or DNA binding is necessary for the dimethylation of K140, we disrupted these properties by using the STAT3 R214/215A and R414/417A mutants (11), expressing them stably in A4 cells at levels comparable to the level of STAT3 in parental DLD1 cells. When STAT3 DNA binding was disrupted by the R414/417A mutation, the level of phosphorylation of STAT3 on S727 was dramatically decreased and little or no STAT3-K140me2 was observed (Fig. 2A), demonstrating that STAT3 DNA binding is required for both the phosphorylation of S727 and for the dimethylation of K140. Similar results were found when STAT3 nuclear translocation was disrupted through the R214/215A mutation (Fig. 2A). Both S727-phosphoryl-STAT3 and STAT3-K140me2 were detected only in nuclei and not in the cytoplasm when these subcellular fractions were examined by Western analysis (Fig. 2B). Finally, when S727 phosphorylation was blocked by the S727A mutation, K140 dimethylation was dramatically decreased (Fig. 2A). From these data we conclude that nuclear entry and DNA binding are necessary for S727 phosphorylation and that both DNA binding and S727 phosphorylation are necessary for K140 dimethylation. Our conclusion that the phosphorylation of STAT3 on S727 can only take place after it is bound to DNA mirrors the similar conclusion of Sadzak et al. (12) for the phosphorylation of STAT1 on S727 in response to IFN.
Fig. 2.
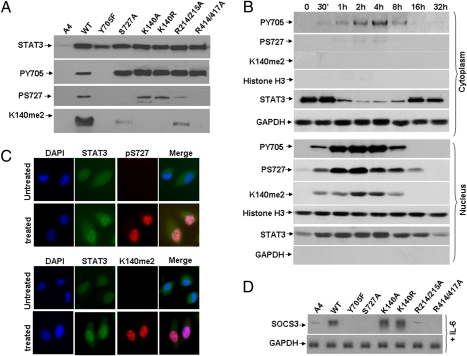
Nuclear import and DNA binding are required for IL-6–induced S727 phosphorylation and K140 dimethylation of STAT3. (A) Western analyses for STAT3, S727-phosphoryl-STAT3, and STAT3-K140me2. STAT3-null A4 cells were infected with retroviral constructs and stable pools were selected with G418. A4, STAT3-null cells; WT, YF, S727A, K140A, K140R, R214/215A and R414/417A, A4 cells expressing normal levels of wild-type or mutant STAT3 proteins. Total lysates prepared from cells treated with IL-6 for 4 h were analyzed by Western blot. (B) A4 cells expressing wild-type STAT3 were treated with IL-6. Cytoplasmic and nuclear fractions representing equal numbers of cells were separated by electrophoresis and analyzed by Western blot. (C) A4 cells expressing wild-type STAT3 were grown on coverslips to 20 to 30% confluence, then treated with IL-6 for 4 h, followed by staining with primary antibodies directed against STAT3, S727-phosphoryl-STAT3 and STAT3-K140me2. Following staining with DAPI (blue nuclear stain) and fluorescent secondary antibodies against STAT3 (green), S727-phosphoryl-STAT3 (red) or STAT3-K140me2 (red), the cells were examined by confocal microscopy. The yellow/pink pixels in the composite image demonstrate the close association of the two proteins. (D) Northern analysis of gene expression. Cells were treated with IL-6 as above for 4 h and total RNAs were analyzed for GAPDH and SOCS3.
In cells treated with IL-6 for various times, S727-phosphoryl-STAT3 was present primarily in the nucleus and was detected at only a very low level in the cytoplasm, whereas STAT3-K140me2 was detected only in the nucleus at all times (Fig. 2B). The maximum levels of Y705-phosphoryl-STAT3 and S727-phosphoryl-STAT3 were observed between 1 and 4 h, whereas the highest level of STAT3-K140me2 were seen somewhat later, between 2 and 4 h (Fig. 2B, nucleus), suggesting that methylation occurs after STAT3 has been phosphorylated on Y705. Analysis by immunofluorescence confirmed that, in untreated cells, STAT3 was distributed similarly between cytoplasm and nuclei, although in IL-6–treated cells STAT3 was seen mainly in nuclei (Fig. 2C, Upper). As expected, no signal for S727-phosphoryl-STAT3 was detected in untreated cells, but in IL-6–treated cells, a strong nuclear signal was present (Fig. 2C). STAT3-K140me2 was not detected in untreated cells, whereas in IL-6–treated cells a strong signal was present, but only in nuclei (Fig. 2C, Lower). Finally, SOCS3 expression induced by IL-6 was higher in cells expressing the K140A or K140R mutants than in cells expressing wild-type STAT3, whereas S727A-, R214/215A-, and R414/417A-STAT3 all failed to induce SOCS3 in response to IL-6 (Fig. 2D).
STAT3 Is Methylated by SET9 and Demethylated by LSD1.
To screen for the enzymes that methylate and demethylate STAT3, flag-tagged human STAT3 was expressed in Escherichia coli, purified by affinity chromatography, and incubated with the purified histone methylases SET9, SMYD2, SET8, G9a, GLP, and SETDB1, and a methyl donor (13). Of the enzymes tested, only SET9 was capable of methylating STAT3 (Fig. S4A). However, only truncated forms of STAT3 and not the full-length protein were methylated in this assay. An explanation is provided by an additional experiment in which STAT3 was purified from A4-STAT3 cells, with or without treatment with IL-6, followed by incubation with SET9 in vitro. Full-length STAT3 from untreated cells was not methylated, but truncated forms generated during the in vitro incubation were modified (Fig. S4B). However, the full-length STAT3 generated following IL-6 treatment, which leads to phosphorylation of Y705, could be methylated readily in vitro, revealing that the K140 methylation site is masked in full-length unphosphorylated STAT3 but becomes accessible to SET9-catalyzed dimethylation following the major conformational change that accompanies STAT3 tyrosine phosphorylation (14, 15).
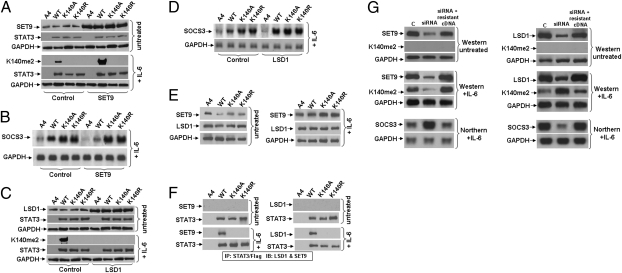
We transfected a construct encoding SET9 into A4 cells expressing wild-type, K140A, or K140R STAT3 (Fig. 3A, Upper). Increased levels of SET9 increased the dimethylation of K140 in response to IL-6 (Fig. 3A, Lower). Consistently, there was a decrease in the expression of SOCS3 in cells expressing wild-type STAT3 (Fig. 3B). As expected, the levels of SOCS3 were not affected by increased levels of SET9 in cells expressing STAT3 K140A or K140R (Fig. 3B). Coimmunoprecipitation experiments showed that SET9 binds to STAT3, but only following treatment of cells with IL-6 (Fig. 3 E and F). Furthermore, knocking the expression of SET9 down with siRNA significantly increased SOCS3 expression (Fig. 3G, Left). We conclude that SET9, which also modifies H3K4, NFκB, and p53 (13), is required for the dimethylation of K140 of STAT3 in response to IL-6. LSD1 demethylates H3K4 and H3K9 (16, 17), p53 (18), and DNMT1 (19). We transfected an expression construct for LSD1 transiently into A4 cells expressing wild-type, K140A, or K140R STAT3 (Fig. 3C, Upper) and found a significant decrease in the level of STAT3-K140me2 (Fig. 3C, Lower). Consistently, demethylation of K140me2 STAT3 increases SOCS3 expression in response to IL-6 (Fig. 3D) and LSD1 knock-down decreases SOCS3 expression in response to IL-6 (Fig. 3G, Right). The results of coimmunoprecipitation experiments showed that LSD1 binds to STAT3, but only following treatment of the cells with IL-6 (Fig. 3 E and F). In addition, LSD1 was able to reverse the methylation of STAT3 K140 in vitro (Fig. S3B). To prove the specificity of the siRNAs complementary to SET9 and LSD1, we showed that they did not cause degradation of SET9 and LSD1 mRNAs in which the target sequence was changed but the protein sequence was not (Fig. 3G).
Fig. 3.
STAT3 is methylated by SET9 and demethylated by LSD1. (A) Cells were transfected transiently with pcDNA3.1-SET9. After 48 h, the cells were treated with IL-6 for 4 h and total cell lysates were analyzed by Western blot. A4, STAT3-null cells; WT, K140A and K140R, A4 cells expressing normal levels of STAT3 proteins. (B) Cells were transfected transiently with pcDNA3.1-SET9. After 48 h, the cells were treated with IL-6 for 4 h and total RNAs were analyzed by Northern blot. (C) Cells were transfected transiently with pcDNA3.1-LSD1. After 48 h, the cells were treated with IL-6 for 4 h and total cell lysates were analyzed by Western blot. A4, STAT3-nullcells; WT, K140A and K140R, A4 cells expressing normal levels of STAT3 proteins. (D) Cells were transfected transiently with pcDNA3.1-LSD1. After 48 h, the cells were treated with IL-6 as above for 4 h and total RNAs were analyzed by Northern blot. (E) Western analyses for SET9 and LSD1. A4 cells were infected with retroviral constructs and stable pools of cells were selected with G418. DLD1, parental cells; A4, STAT3-null cells; WT, K140A, and K140R, A4 cells expressing normal levels of STAT3 proteins. The cells were treated with IL-6 for 4 h and total cell lysates were analyzed by Western blot. (F) STAT3 was immunoprecipitated from whole-cell extracts of the above cells by using anti-Flag M2 beads. Western analyses were performed to detect SET9 and LSD1. (G) A4 cells expressing wild-type STAT3 were transfected transiently with siRNAs directed against SET9 or LSD1 and, 24 h later, the cells were transfected again with the same siRNAs. After an additional 24 h, some of the cells were transfected with siRNA-resistant SET9 or LSD1 cDNAs. After another 24 h, some of the cells were treated with IL-6 for 4 h more and analyzed for SOCS3 expression by Northern blot and some were left untreated and analyzed for SET9 and LSD1 expression by Western blot.
Comparison of Genes Induced by IL-6 in Wild-Type Cells and in A4 Cells Expressing K140R STAT3.
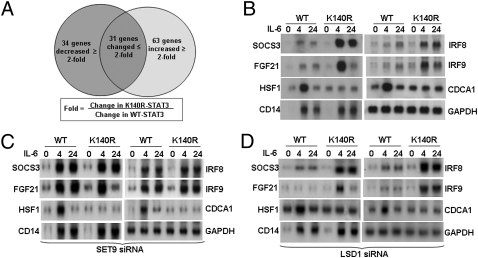
Cells expressing wild-type or K140R STAT3 were treated with IL-6 for 4 h. Among 128 IL-6–responsive mRNAs whose levels differed by twofold or more in the treated versus untreated samples, the expression of 97 changed by twofold or more in the treated K140R samples; 63 were increased and 34 were decreased (Fig. 4A and Table S1). SOCS3, FGF21, IRF8, and IRF9 represent mRNAs whose expression is increased in K140R STAT3 cells, HSF1 and CDCA1 represent mRNAs with decreased expression, and CD14 represents mRNAs induced similarly in both cells. Northern data for these mRNAs (Fig. 4B) confirm the array results, revealing three classes of genes that respond differently to disruption of STAT3 methylation at K140. To be sure that these expression patterns are specific for STAT3, the levels of SET9 and LSD1 mRNAs were knocked down with siRNAs. As shown in Fig. 4C, reduced expression of SET9 increased the expression of the mRNAs that are down-regulated by the dimethylation of K140 (i.e., SOCS3, IRF8, and IRF9), and reduced expression of LSD1 decreased the expression of these mRNAs (Fig. 4D). However, the expression of mRNAs that were not affected by STAT3 dimethylation (i.e., HSF1 and CD14) also were unaffected by changes in the expression of SET9 or LSD1 (Figs. 4 C and D).
Fig. 4.
Comparison of genes induced by IL-6 in wild-type cells or in A4 cells expressing K140R STAT3. (A) Cells were treated with IL-6 for 4 or 24 h or were untreated. Total RNAs were analyzed by using an Illumina gene array. Messenger RNAs showing a change in expression of twofold or more, compared with their expression in untreated cells, were scored. Fold-change was calculated from the ratio (K140R-STAT3 treated for 4 h/untreated) ÷ (WT-STAT3 treated for 4 h/untreated). (B) Northern analysis of gene expression. Total RNAs were used and GAPDH was analyzed on the same transfers. (C) SET9 siRNA was used as described in Fig. 3G. After 48 h more, the cells were treated with IL-6 for 4 or 24 h. Total RNA was extracted for Northern analysis. (D) LSD1 siRNA was used as described in Fig. 3G. After an additional 48 h, the cells were treated with IL-6 for 4 or 24 h. Total RNA was extracted for Northern analysis.
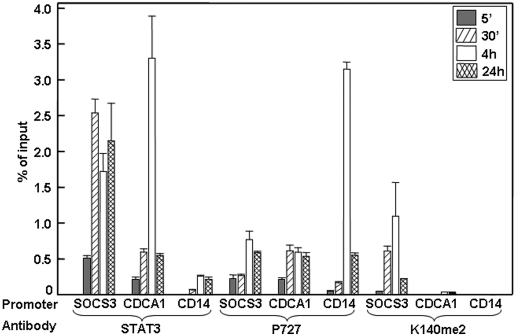
STAT3-K140me2 Binds to the SOCS3 Promoter Selectively.
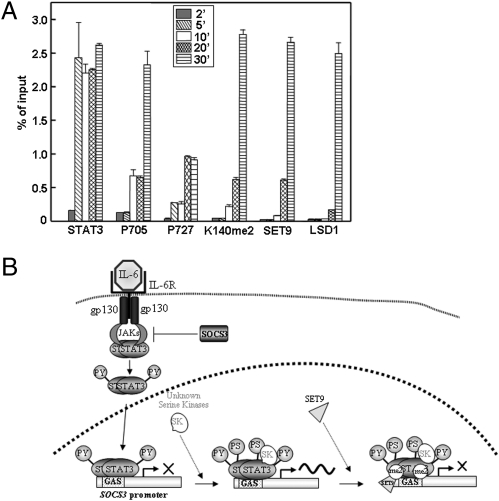
To test the promoter occupancy of different forms of STAT3, ChIP experiments were performed with IL-6–treated cells (Fig. 5). Total STAT3 and S727-phosphoryl-STAT3 are detected on examples of all three classes of promoters (SOCS3, CDCA1, and CD14), whereas K140me2 STAT3 is detected on the SOCS3 promoter but not on the CDCA1 or CD14 promoters. The SOCS3 promoter was then analyzed to determine the sequence of events. The data shown in Fig. 6A, obtained by qPCR analysis, represent quantitatively the kinetics of appearance of each species on the SOCS3 promoter. Interestingly, in response to IL-6, the binding of Y705-phosphoryl-STAT3 to the promoter can be detected by 2 min. S727-phosphoryl-STAT3 is barely detectable at 2 min and increases at later times. STAT3-K140me2 and SET9 are observed on the promoter at the same times, later than Y705- and S727-phosphoryl-STAT3, and LSD1 binds last. These results are pictured in Fig. 6B, except that the events including and following the binding of LSD1 are not represented because we do not yet understand the connection between the methylation of STAT3 and the dephosphorylation of Y705 well enough to represent it in a model.
Fig. 5.
STAT3-K140me2 binds to the SOCS3 promoter selectively. A ChIP assay was performed with chromatin from IL-6–treated or untreated A4 cells expressing wild-type STAT3, using antibodies against total STAT3, S727-phosphoryl-STAT3, or STAT3-K140me2. The immunoprecipitated DNA was amplified by qPCR, using primers specific for SOCS3, CDCA1, and CD14. Values (percent of the inputs) are the mean ± SD of triplicates performed on three separate chromatin preparations. Data were analyzed using origin75 software and compared by using the unpaired Student's t test. All of the values obtained with each antibody were corrected for background by subtracting the value for untreated cells, which was about 0.1% of the input in each case.
Fig. 6.
The sequence of events that follows the binding of STAT3 homodimers to the SOCS3 promoter. (A) CHIP assays. Quantitative ChIP assays were performed with chromatin from IL-6–treated or untreated A4 cells expressing wild-type STAT3, using antibodies against total STAT3, Y705-phosphoryl-STAT3, S727-phosphoryl-STAT3, STAT3-K140me2, SET9, and LSD1. The immunoprecipitated DNA was amplified by qPCR, using primers specific for SOCS3. Values (percent of the inputs) are the mean ± SD of triplicates performed on three separate chromatin preparations. Data were analyzed using origin75 software and compared by using the unpaired Student's t test. All of the values obtained with each antibody were corrected for background by subtracting the value for untreated cells, which was about 0.1% of the input in each case. Note that the data shown are averages, representing mixed populations of cells. We expect that each individual promoter will not be able to bind both the methyltransferase and the demethylase simultaneously. (B) A model for the sequence of events that follows the binding of STAT3 homodimers to the SOCS3 promoter. Events following the binding of LSD1 and demethylation of STAT3 are not represented because they are not well understood at present.
Discussion
Enzymatic modification of histones by methylation, acetylation, and phosphorylation helps to change chromatin structure in response to the binding of transcriptional activators and repressors (20, 21). We now present the unique observation that a transcription factor can be modified when it is bound to specific promoters, by the same enzymes that modify histones, with major functional consequences. This observation is important for interpreting experiments in which histone-modifying enzymes have been manipulated or have been mutated, as the modification of not only histones but also of other promoter-bound proteins may be affected. In addition to STAT3, p53 is modified on three different lysine residues by histone methyl transferases and demethylases (3, 18, 22), and there is recent evidence for similar modifications, also catalyzed by histone methyl transferases and demethylases, of at least three lysine residues of NFκB. K37 of the p65 subunit is monomethylated by SET9 (4), and K218 and K221 are monomethylated and dimethylated, respectively, by NSD1 and demethylated by FBXL11 (5). All these modifications activate NFκB function. For p53, the reactions occur on K370, K372, and K382, with consequences for function that depend on the site and the degree of methylation (3, 18, 22–24). K370 is monomethylated by the H3K4 methyl transferase SMYD2, repressing transcription, and is dimethylated at the same site by an unknown methyl transferase. Dimethylation activates p53 by providing a binding site for the coactivator 53BP1. Because the phosphorylation of S727 of STAT3 seems to be a prerequisite for the dimethylation of K140, it is possible that phosphoryl-S727 helps to provide a binding site for the lysine methyltransferase SET9. We do not know how dimethylation of K140 inhibits the binding of STAT3 to promoters. However, by analogy with the above examples, a possibility is that the binding of an accessory protein to this dimethyllysine residue of STAT3 facilitates its dissociation. Although SET9 is necessary for the dimethylation of K140 of STAT3 in cells, it is possible that it may not be sufficient, because its ability to transfer a second methyl group is controversial. However, some reports do show that SET9 is capable of dimethylating substrates (22, 25–27), and our own data show that it can dimethylate K140 of STAT3 in vitro (Fig. S4).
Our data provide strong support for the conclusion that STAT3 is methylated in the nucleus and not in the cytosol. All reported lysine methylations and demethylations of transcription factors, including STAT3, are carried out by histone-modifying enzymes, which are chromatin-bound nuclear proteins. STAT3 is not methylated if mutations prevent it from being transported into the nucleus or binding to DNA, even though the corresponding STAT3 phosphotyrosine dimers are formed (Fig. 2A). STAT3-K140me2 is found only in the nucleus (Fig. 2B). Additional data show that STAT3 methylation takes place on chromatin and not in the nucleoplasm. Kinetic ChIP experiments reveal the sequence of events on the STAT3-regulated SOCS3 promoter (Fig. 6A). The sequential binding of tyrosine-phosphorylated STAT3 dimers, followed by serine 727 phosphorylation, binding of the methylase and lysine methylation, and binding of the demethylase, is very hard to reconcile with a model in which the various forms of STAT3 come off the promoter to be modified in the nucleoplasm and then rebind. Further strong evidence comes from the independent finding that the phosphorylation of S727 of STAT3 is a prerequisite for the methylation of lysine 140 (Fig. 2A), consistent with the kinetics that show serine phosphorylation somewhat earlier than lysine methylation (Fig. 6A). Finally, selective methylation of STAT3 only on a subset of STAT3-induced promoters (Figs. 4 and 5) again argues against a model in which modification in the nucleoplasm precedes the binding of STAT3 to promoters.
Major findings of this work are that STAT3 is reversibly dimethylated on K140 by histone-modifying enzymes only when it is part of a promoter-bound complex and that this modification affects only subsets of STAT3-activated genes. The ability to control the methylation of a transcription factor in a promoter-specific manner provides a way to modulate the time of residence of the transcription factor independently on each promoter (by modulating affinity) and the strength of the transactivating signal. Thus, a single modified species of an activated transcription factor can regulate the expression of separate subclasses of promoters differentially. The H3K4 methyl transferase SET9 is recruited to the SOCS3 promoter only after STAT3, providing a potential mechanism for signal-dependent induction of an activating histone mark. Modifications of promoter-bound transcription factors are likely to provide docking sites for proteins that carry out important functions. Enzymes that modify histones and transcription factors may also modify other promoter-bound proteins and, thus, additional substrates of histone lysine methyl transferases and demethylases may be revealed by future experiments.
Methods
Antibodies against STAT3K140me2 were produced at Lanzhou University by immunizing rabbits. Antibodies were purified by affinity chromatography, and negative screening strategies were applied to remove antibodies that cross-reacted with wild-type STAT3. STAT3-null human colon cancer A4 cells were generated by a homologous recombination knockout strategy at Case Western Reserve University. For all other details concerning cells, reagents, and experimental procedures, see SI Methods.
Supplementary Material
Acknowledgments
We thank Dr. Judith Drazba of the Cleveland Clinic for expert help with confocal microscopy. This work was supported by Grants P01 CA62220 (to G.R.S.), R01 CA127590 (to Z.W.), NCET-08-0260 from the Ministry of Education and 2009DFA30990 from the Ministry of Science and Technology of the People's Republic of China, and 0708WCGA149 from the Gansu Provincial Ministry of Science and Technology (to J.Y.). The mass spectrometry experiments were supported in part by funds from the Case Comprehensive Cancer Center.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016147107/-/DCSupplemental.
References
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Ea CK, Baltimore D. Regulation of NF-kappaB activity through lysine monomethylation of p65. Proc Natl Acad Sci USA. 2009;106:18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu T, et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA. 2010;107(1):46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akira S, et al. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77(1):63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 7.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 8.O'Shea JJ, Kanno Y, Chen X, Levy DE. Cell signaling. Stat acetylation—A key facet of cytokine signaling? Science. 2005;307:217–218. doi: 10.1126/science.1108164. [DOI] [PubMed] [Google Scholar]
- 9.Ray S, Boldogh I, Brasier AR. STAT3 NH2-terminal acetylation is activated by the hepatic acute-phase response and required for IL-6 induction of angiotensinogen. Gastroenterology. 2005;129:1616–1632. doi: 10.1053/j.gastro.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 10.McKendry R, et al. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc Natl Acad Sci USA. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma J, Zhang T, Novotny-Diermayr V, Tan AL, Cao X. A novel sequence in the coiled-coil domain of Stat3 essential for its nuclear translocation. J Biol Chem. 2003;278:29252–29260. doi: 10.1074/jbc.M304196200. [DOI] [PubMed] [Google Scholar]
- 12.Sadzak I, et al. Recruitment of Stat1 to chromatin is required for interferon-induced serine phosphorylation of Stat1 transactivation domain. Proc Natl Acad Sci USA. 2008;105:8944–8949. doi: 10.1073/pnas.0801794105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chuikov S, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 14.Becker S, Groner B, Müller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394(6689):145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 15.Braunstein J, Brutsaert S, Olson R, Schindler C. STATs dimerize in the absence of phosphorylation. J Biol Chem. 2003;278:34133–34140. doi: 10.1074/jbc.M304531200. [DOI] [PubMed] [Google Scholar]
- 16.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449(7158):105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 20.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 23.Ivanov GS, et al. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol. 2007;27:6756–6769. doi: 10.1128/MCB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi X, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon T, et al. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. EMBO J. 2003;22:292–303. doi: 10.1093/emboj/cdg025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakrabarti SK, Francis J, Ziesmann SM, Garmey JC, Mirmira RG. Covalent histone modifications underlie the developmental regulation of insulin gene transcription in pancreatic beta cells. J Biol Chem. 2003;278:23617–23623. doi: 10.1074/jbc.M303423200. [DOI] [PubMed] [Google Scholar]
- 27.Francis J, Chakrabarti SK, Garmey JC, Mirmira RG. Pdx-1 links histone H3-Lys-4 methylation to RNA polymerase II elongation during activation of insulin transcription. J Biol Chem. 2005;280:36244–36253. doi: 10.1074/jbc.M505741200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.