Abstract
Nucleotide variations, including SNPs, in the coding regions of disease genes are important targets for RNAi treatment, which is a promising medical treatment for intractable diseases such as triplet repeat diseases. However, the identification of such nucleotide variations and the design of siRNAs conferring disease allele-specific RNAi are quite difficult. In this study we developed a pull-down method to rapidly identify coding SNP (cSNP) haplotypes of triple repeat, disease-causing alleles, and we demonstrated disease allele-specific RNAi that targeted cSNP sites in mutant Huntingtin alleles, each of which possessed a different cSNP haplotype. Therefore, the methods presented here allow for allele-specific RNAi knockdown against disease-causing alleles by using siRNAs specific to disease-linked cSNP haplotypes, and advanced progress toward tailor-made RNAi treatments for triplet repeat diseases.
Keywords: disease allele-specific silencing, Huntington disease
Suppression of disease-causing alleles or elimination of the mutant gene products is a potential approach to treatment of intractable diseases caused by dominate-negative alleles, such as triplet repeat diseases (1–3). For these treatments to be side-effect free, expression of the functional wild-type alleles and gene products must be unaffected. RNAi may be an applicable tool in such medical treatment; however, these treatments will require atypical RNAi silencing that is capable of discriminating between target (mutant) and nontarget (wild-type) alleles. The design and selection of siRNAs conferring allele-specific RNAi (ASP-RNAi) (4–8) are vital, but they are difficult. In the case of triplet repeat diseases, the expanded trinucleotide repeats of triplet repeat disease-causing alleles are not suitable RNAi targets for ASP-RNAi, because identical repeat sequences are present in corresponding normal alleles. Therefore, identification of nucleotide variations that are suitable targets for ASP-RNAi in the disease-causing alleles (9, 10) is required; however, it is also difficult and time-consuming. Accelerating the identification of useful nucleotide variations is essential to ASP-RNAi against disease-causing alleles, or disease-causing allele-specific RNAi (DIAS-RNAi), particularly for triplet repeat diseases.
Here we report an innovative procedure that facilitated allele-specific silencing of disease-causing alleles in Huntington disease (HD), a triplet repeat disease. We developed an easy-to-use method, the pull-down method, for identification of the nucleotide variations, including coding SNPs (cSNPs), that were linked with aberrantly expanded trinucleotide repeats in the disease-causing alleles. In addition, this method greatly reduced the processing time necessary to identify the disease-causing alleles. Together with our in vitro assay system for assessment of siRNAs conferring ASP-RNAi [described previously (4, 6, 8)], we achieved DIAS-RNAi knockdown against mutant Huntingtin (HTT) alleles with selected siRNAs that targeted cSNP sites within mutant alleles that were determined by the pull-down method.
Results
Biased Recovery of Triplet Repeat Disease-Causing Alleles by Pull-Down Method.
We investigated HTT, which is the responsible gene for HD, a typical triplet repeat disease. Whereas the normal HTT alleles possess ≈18 repeats of CAG trinucleotide in exon 1, the mutant HTT alleles carry aberrantly expanded CAG trinucleotide repeats (>36 repeats; average, 42 repeats) (11) that result in expansion of the polyglutamine tract in the HTT protein. These aberrant, expanded polyglutamine tracts in mutant HTT proteins are closely related to the onset and severity of HD (11–13).
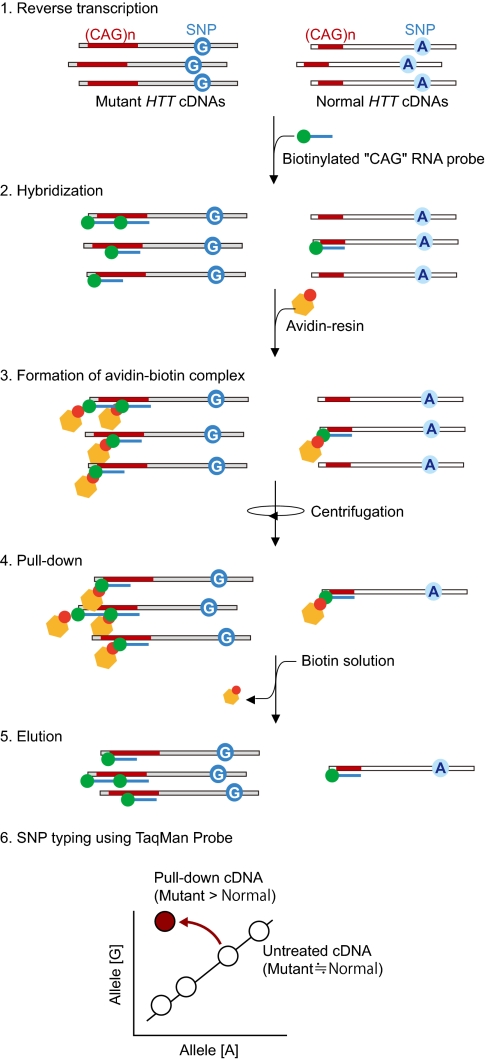
To clone and characterize the mutant HTT alleles in HD patients, we developed a method, the pull-down method (Fig. 1; detailed in Materials and Methods), that preferentially recovered mutant HTT alleles relative to the normal HTT allele. The method was based on the nucleic acid hybridization and fractionation techniques. Briefly, cDNAs were synthesized by reverse transcription using total RNA extracted from HD patients’ specimens as template; the cDNA was subjected to hybridization with the biotinylated CAG ribonucleotide repeat probe (biotin-CAG RNA probe) in vitro. The use of RNA as a probe was crucial for the success of subsequent assays based on PCR. This is because DNA probes, but not RNA probes, associated with cDNA naturally worked as contaminating PCR primers, causing unwanted PCR amplification in subsequent assays. Therefore, RNA probes, unlike DNA probes, do not interfere with PCR in subsequent assays. The hybridized cDNAs were collected by pull-down with avidin-resin, and nucleotide variations (cSNPs and CAG repeats) in disease alleles of interest were examined by the TaqMan SNP Genotyping assay (Applied Biosystems) and PCR analyses (detailed in Materials and Methods). Importantly, DNA amplification by PCR or by cloning with bacteria was not required for the pull-down step; therefore, the first synthesized cDNAs were free from any artificial nucleotide changes involving DNA amplification.
Fig. 1.
Schematic view of the pull-down method. cDNAs prepared from patients’ RNAs (1) were subjected to hybridization with the biotin-CAG RNA probe (2); and the hybridized cDNAs were recovered by pull-down with avidin-resin (3, 4), followed by elution (5). The recovered cDNAs were examined by the TaqMan SNP Genotyping assay (6). When SNP sites were heterozygous, the SNP typing with the recovered cDNAs plotted the spots distant from the standard curves.
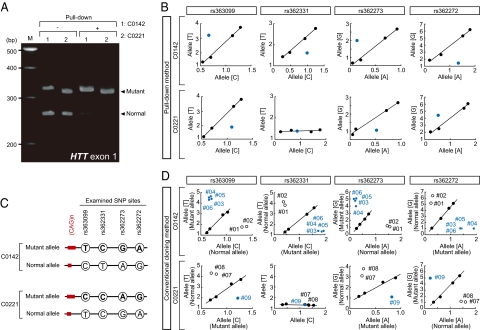
Fig. 2 shows the results of the typing with the cDNAs recovered by the pull-down method. The pull-down method resulted in biased recovery of the mutant HTT cDNAs; cDNA samples subjected to pull-down retained the mutant allele but not the wild-type allele (Fig. 2A). The subsequent TaqMan SNP Genotyping assays of the pulled-down cDNAs exhibited the spots (blue spots, Fig. 2B) plotted away from the standard curves at heterozygous cSNP sites; therefore, nucleotides at the cSNP sites and the corresponding cSNP haplotypes in mutant HTT cDNAs were readily determined (Fig. 2C). In contrast, spots from a homozygous cSNP site (e.g., the rs362331 SNP in C0221) fell on the standard curve (Fig. 2B). Taken together, our data demonstrate that we successfully extracted disease-causing HTT alleles with the pull-down method, and we determined cSNP haplotypes linked with the aberrant, expanded CAG trinucleotide repeats. We further demonstrated that the pull-down method also allowed for recovery and examination of disease-causing HTT cDNAs directly prepared from peripheral blood samples of HD patients (Fig. S1), suggesting that the method is applicable in rapid diagnosis and routine medical examinations.
Fig. 2.
Biased recovery of disease-causing HTT alleles. (A) cDNAs prepared from two HD cases (C0142 and C0221) were subjected to the pull-down method (+), and then the length of the CAG trinucleotide repeat array in HTT was investigated by PCR analysis. Untreated cDNAs (−) were also examined as a control. (B) cDNAs (C0142 and C0221), which were the same as in A, were subjected to the TaqMan SNP Genotyping assay, and the results were plotted in blue. Standard curves (indicated in black) were constructed by the same SNP typing using serially diluted (0.01, 1, 10 ng) untreated cDNAs. The examined coding SNP (cSNP) sites are indicated. x- and y-axes indicate relative fluorescent intensity obtained from the TaqMan probes against the indicated alleles, respectively, and allelic nucleotides at the cSNP sites are also shown in parentheses. (C) Linkage between the expanded CAG trinucleotide repeats and cSNPs was estimated in each HD case according to the results of the cSNP typing in B. (D) Confirmatory experiments by conventional methods. The same SNP typing as in B was carried out using the isolated plasmids carrying the HTT alleles, which were obtained by conventional methods (Fig. S2). Dots in blue and open circles represent the results of the plasmids containing the mutant and normal HTT alleles, respectively. Indicated clone (plasmid) numbers correspond to those indicated in Fig. S2.
The reliability of the data described above was confirmed with conventional PCR-based cloning of the entire ORFs of mutant and normal HTT alleles from patients C0142 and C0221 (Fig. S2), followed by determination of nucleotides at the cSNP sites and of the length of the CAG trinucleotide repeats (Fig. 2D). The confirmatory experiments further highlighted that, whereas the conventional methods took several days (7–14 d) to determine the linkage between the cSNPs and CAG repeat in the disease-causing alleles (almost the same as in ref. 9), our current procedure required ≈6 h after isolation of total RNAs from HD patient's specimens (Table S1). Therefore, the pull-down method greatly reduced the processing time.
Versatility and Capability of the Pull-Down Method.
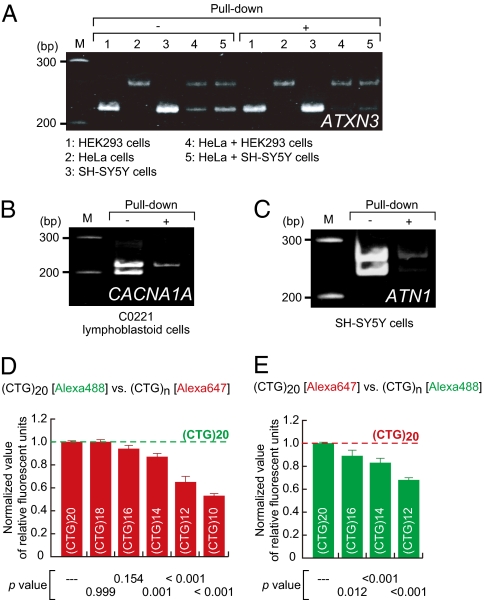
Other triplet repeat disease genes carrying CAG trinucleotide repeats were examined to assess versatility of the pull-down method. As shown in Fig. 3 A–C, the pull-down method preferentially recovered cDNAs of the alleles with longer CTG trinucleotide repeats (complementary CAG trinucleotide repeats) of three genes— ATXN3, CACNA1A, and ATN1—associated with three different triplet repeat diseases—spinocerebellar ataxia 3, spinocerebellar ataxia 6, and dentatorubral-pallidoluysian atrophy, respectively. Therefore, the data indicated that the method could be applied to identify disease-causing alleles in other triplet repeat diseases.
Fig. 3.
Pull-down of (A) ATXN3, (B) CACNA1A, and (C) ATN1 cDNAs carrying longer CAG trinucleotide repeats. cDNAs were prepared from indicated human cell lines and treated by the pull-down method. The recovered cDNAs were examined by PCR analysis and assessed CAG trinucleotide repeat array length in each gene as in Fig. 2A. Because the CAG trinucleotide repeat arrays in ATXN3 (A) were homozygous in all of the samples investigated, mixtures of the HEK293 and HeLa cDNAs (Sample 4) and of the HeLa and SH-SY5Y cDNAs (Sample 5) were prepared and then examined. (D and E) Allele discrimination capability of the pull-down method was examined. The Alexa 488- and Alexa 647-labeled CTG oligonucleotides with various numbers of the CTG trinucleotide repeats, which are complementary to the biotin-CAG RNA probe, were used as targets. The number (n) of the CTG repeats is indicated as (CTG)n. (CTG)20 oligonucleotides, the longest CTG repeats arrays (20 repeats), were labeled with Alexa 488 (D) or Alexa 647 (E) and individually mixed with a competitor, a (CTG)n oligonucleotide labeled with the other fluorescent dye; these mixtures were then subjected to the pull-down assay. The fluorescence intensities of the oligonucleotides recovered by the pull-down method were examined. Ratios of normalized fluorescent intensities of the competitors to that of the (CTG)20 oligonucleotide are shown. Data are averages of six independent tests. Error bars indicate SD. P values were calculated by Dunnett's multiple comparison test.
We further investigated the allele discrimination capability of the method using Alexa 488- and Alexa 647-labeled CTG oligonucleotides with various numbers of the CTG trinucleotide repeats (Fig. 3 D and E). Oligonucleotide with 20 CTG repeats [(CTG)20; Table S2] were preferentially recovered when CTG oligonucleotides with 16 CTG repeats were used as competitors. Therefore, the method discriminated between oligonucleotides that differed by only four trinucleotide repeats and may discriminate between actual alleles that differ by at least four trinucleotide repeats.
Allele-Specific Knockdown Against Disease-Causing HTT Alleles by ASP-RNAi Targeting cSNP Sites.
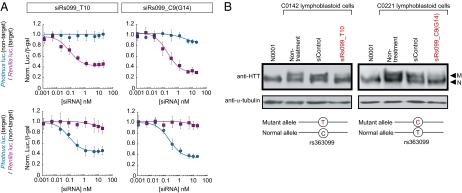
Combining the pull-down method with subsequent cSNP typing and ASP-RNAi silencing may be the foundation for ASP-RNAi treatment for intractable triplet repeat diseases. To test this possibility, we performed ASP-RNAi knockdown against endogenous mutant HTT alleles in lymphoblastoid cell lines established from peripheral blood cells of HD patients. Genotyping of the HTT cSNPs (rs363099, rs362331, rs362273, and rs362272) in six HD cases revealed that three cases were heterozygous (Fig. S3 and Table S3); and the subsequent pull-down method, followed by cSNP typing, determined the cSNP haplotypes associated with the disease-causing (expanded) CAG trinucleotide repeats (Fig. 2C). Notably, the disease-causing HTT alleles differed from one another in their cSNP haplotypes. This suggests that the disease-causing alleles may have different cSNP haplotypes linked with the expanded CAG trinucleotide repeats. Consequently, the therapeutic siRNAs must be designed to target the disease-linked cSNP haplotypes for disease allele-specific RNAi knockdown. We designed and assessed siRNAs targeting the above cSNP sites by means of our in vitro assay system, described previously (4), and competent siRNA(s) in response to each allelic nucleotide at the four cSNP sites was successfully designed (Fig. 4A and Fig. S4–S6; designed siRNAs in Tables S4–S6). The results of Fig. 4A indicated that the designed siRNAs conferred dose-dependent, ASP-RNAi knockdown.
Fig. 4.
Disease allele-specific RNAi knockdown against the mutant HTT alleles carrying distinct cSNP haplotypes. (A) ASP-RNAi knockdown by siRs099_T10 and siRs099_C9(G14) siRNAs, which targeted the T and C alleles, respectively, at the rs363099 cSNP site in HTT were examined by IC50 analysis: the effects of the designed siRNAs on suppression of target alleles and allele discrimination were investigated. Data are the average of four independent determinations. Error bars represent SDs. (B) Western blot analyses of the endogenous HTT protein. On the basis of the C0142 and C0221 cSNP haplotypes (Fig. 2C), appropriate siRNA targeting the rs363099 cSNP site in the respective mutant HTT alleles was chosen and introduced into lymphoblastoid cells from the corresponding HD patient. After 3-d incubation, cell lysate was prepared and examined by Western blotting with anti-human HTT antibody; anti-α-tubulin antibody was used as an internal loading control. The siRNAs and lymphoblastoid cell lines are indicated above each gel. A healthy individual (N0001) was also examined as a control. The mutant and normal HTT isoforms are indicated by M and N, respectively.
On the basis of the cSNP haplotypes of the disease-causing HTT alleles (Fig. 2C), we chose specific siRNAs that targeted the two different disease-causing alleles in HD cases C0142 and C0221 and introduced these siRNAs into HD lymphoblastoid cells derived from cases C0142 and C0221 by electroporation. The effects of the siRNAs on the expression of mutant and wild-type (normal) HTT proteins were examined by Western blotting. We modified the SDS/PAGE method to obtain high resolution of high-molecular-mass polypeptides, such as HTT (detailed in Materials and Methods; Fig. S7). As shown in Fig. 4B and Fig. S8, the levels of the mutant HTT proteins were markedly decreased relative to those of wild-type HTT in the presence of the allele-specific siRNAs, demonstrating that allele-specific silencing of the disease-causing allele by RNAi occurred. Importantly, the disease-causing HTT alleles in C0142 and C0221 possessed different cSNP haplotype; nevertheless, the allele-specific silencing of the mutant allele was achieved with the specific siRNAs targeted to the cSNP haplotypes of the disease-causing allele in individual patients. Thus, the evidence was consistent with the possibility of tailor-made, personalized RNAi knockdown.
Discussion
To realize a side-effect-free RNAi treatment for diseases caused by dominant-negative mutations (e.g., triplet repeat diseases), ASP-RNAi against disease-causing alleles, or DIAS-RNAi, is a useful method. To achieve DIAS-RNAi treatments for triplet repeat diseases, identification of nucleotide variations (e.g., cSNPs) that are linked to aberrant, expanded trinucleotide repeats and are suitable as RNAi targets is absolutely necessary, but it is very difficult. Our strategy, which comprised the pull-down method, cSNP typing, and assessment of siRNAs conferring ASP-RNAi, substantially mitigated these difficulties; and the methods allowed for rapid identification of disease-causing alleles in triplet repeat diseases and rapid screening of competent siRNAs that conferred DIAS-RNAi.
The pull-down method presented in this study was quick and easy, and it was a highly sensitive method; specifically, it discriminated differences as small as four trinucleotide repeats between two oligonucleotides (Fig. 3) and seemed to work for multiple disease-causing repeat sequences simply by using RNA probes complementary to aberrantly expanded repeat sequences. Furthermore, the method substantially accelerated the identification of useful nucleotide variations in disease-causing alleles (Table S1). Taken together, our results presented here suggest that the immediate application of this method to multiple repeat diseases may be feasible.
In this study, we designed competent siRNAs that conferred ASP-RNAi at four cSNP sites in the HTT gene. Previous studies suggested that a set of siRNAs that targeted four different SNP sites could be used to treat ≈80% of all HD patients (14, 15); the siRNAs used in this study may have such potential. The most important point to note is that our procedure successfully achieved ASP-RNAi knockdown of two endogenous disease-causing HTT alleles with different cSNP haplotypes using siRNAs that were specific for each cSNP haplotype (Fig. 4), demonstrating tailor-made RNAi knockdown.
ASP-RNAi that targets allelic nucleotides at cSNP sites, or tailor-made RNAi knockdown, may be applied to many heritable, intractable diseases (e.g., triplet repeat diseases); and it may be readily achievable by our current procedures. Therefore, the methodologies presented in this study may pave the way to treatment of currently intractable triplet repeat diseases, and they greatly contribute to the achievement of tailor-made RNAi treatment as a next-generation RNAi therapy.
Materials and Methods
Pull-Down Method.
Total RNA was extracted from patients’ peripheral blood samples and cultured cells and was used as template for cDNA synthesis using oligo(dT) primers and reverse transcriptase. A 100-ng sample of cDNA was mixed with 100 pmol of the biotinylated CAG ribonucleotide repeat probe, 5′-(biotin)–CAGCAGCAGCAGCAGCAGCA-3′, in 40 μL hybridization buffer [10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 400 mM sodium chloride, and 30 mM sodium citrate], and this mixture was heat-denatured at 95 °C for 5 min followed by annealing at room temperature for more than 30 min. After annealing, 10 μL of SoftLink Soft Release Avidin Resin (Promega) pretreated at 42 °C for over 1 h in blocking buffer [10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 100 mM sodium chloride, 1% (w/vol) BSA, and 1 mg/mL yeast RNA] was added, and blocking buffer was added to a final volume of 400 μL. The mixture was incubated at 37 °C for 30 min with gentle shaking. After incubation, the biotin–avidin complexes were collected by centrifugation at 1,500 × g for 1 min, and the complexes were washed three times with wash buffer [10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 100 mM sodium chloride]. The bound cDNAs were eluted from the complexes with 10 μL of 5 mM biotin solution at room temperature for 10 min and collected by centrifugation at 1,500 × g for 1 min. The recovered cDNAs were examined by PCR analyses and SNP typing (described below).
Cell Culture.
HeLa and HEK293 cells were grown at 37 °C in DMEM [Wako Pure Chemical Industries (Wako)] supplemented with 10% FBS (Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin (Wako) in a 5% CO2-humidified chamber. SH-SY5Y cells were grown at 37 °C in DMEM: Nutrient Mixture F-12 (DMEM/F-12, 1:1) medium (Invitrogen) supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin (Wako). Epstein-Barr virus-transformed human lymphoblastoid cell lines were cultured at 37 °C in RPMI 1640 medium (Sigma) supplemented with 10% FBS (Japan Bio Serum), 110 mg/L sodium pyruvate (Wako), 4,500 mg/L d-glucose (Wako), 100 U/mL penicillin, and 100 μg/mL streptomycin (GIBCO) in a 5% CO2 humidified chamber.
RNA and DNA Oligonucleotides.
DNA oligonucleotides and siRNAs used in this study were synthesized by and purchased from Sigma-Aldrich. The biotinylated CAG ribonucleotide repeat probe was also obtained from Sigma-Aldrich.
Total RNA Preparation and cDNA Synthesis.
Total RNAs were extracted from cultured human cells using TRIzol Reagent (Invitrogen) and subjected to cDNA synthesis using Oligo(dT)15 primers (Promega) and a SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions.
PCR Analyses.
PCR analysis was carried out using the primer sets described below and a PrimeSTAR HS DNA polymerase with GC buffer (TaKaRa Bio), according to the manufacturer's instructions. The GeneAmp PCR system 9700 (Applied Biosystems) was used as the thermal cycler, and the thermal cycling profiles were as follows: heat denaturation at 95 °C for 5 min, 26–36 cycles of amplification including denaturation at 98 °C for 10 s, annealing at 60 °C for 5 s, and extension at 72 °C for 30 s The resultant PCR products were electrophoretically separated in 5% polyacrylamide gels and visualized by ethidium bromide staining.
The sequences of the PCR primers used were as follows:
Huntingtin (HTT) exon1 primer set:
5′-ATGGCGACCCTGGAAAAGCTGA-3′
5′-GGTCGGTGCAGCGGCTCCTC-3′
Huntingtin (HTT) exon1-2 primer set:
5′-ATGGCGACCCTGGAAAAGCTGA-3′
5′-CTGACAGACTGTGCCACTATGTT-3′
Ataxin3 (ATXN3) primer set:
5′-GTAGTTCCAGAAACATATCTCAAGA-3′
5′-CTAGATCACTCCCAAGTGCTCC-3′
Atrophin1 (ATN1) primer set:
5′-CCTCTTAGCCAACAGCAATGCC-3′
5′-GGAGGGAGACATGGCGTAAGG-3′
Voltage-dependent P/Q-type calcium channel subunit alpha-1A (CACNA1A) primer set:
5′-GTGTCCTATTCCCCTGTGATCC-3′
5′-CTCTCCATCCTGGGCGAGCG-3′
SNP Typing.
The rs363099, rs362331, rs362273, and rs362272 SNPs in the HTT coding region were investigated by means of the AB 7300 Real Time PCR System (Applied Biosystems) with a TaqMan Universal PCR Master Mix together with TaqMan SNP Genotyping assays (Applied Biosystems) according to the manufacturer's instructions. In the case of SNP typing of the cDNAs recovered by the pull-down method, samples of nontreated, control cDNAs (10, 1, and 0.01 ng) were used to construct the standard curve.
Electroporation.
The introduction of siRNAs conferring ASP-RNAi into lymphoblastoid cells was carried out by means of the Nucleofector system (Amaxa) according to the manufacturer's instructions. Briefly, the cells (1 × 106 cells per transfection) were suspended in 100 μL of electroporation buffer (Cell Line Nucleofector Solution V; Amaxa) containing 5 μM of each siRNA and then subjected to electroporation with the U-005 pulsing program. After electroporation, the cells were immediately transferred into six-well culture plates containing prewarmed culture medium and incubated for 3 d.
Modified Polyacrylamide Gels.
A vertical slab gel (glass plate: 160-mm long, 160-mm wide, and 1-mm thick) electrophoresis apparatus was used, and the gel formulation was as follows:
For separation gels: 5–20% (step-gradient) polyacrylamide gel (acrylamide/N, N’-methylenebisacrilamide ratio 30:0.135) was prepared in 380 mM Tris-acetate (pH 8.8) containing 0.1% SDS.
For stacking gels: 4% polyacrylamide gel (acrylamide/N, N’-methylenebisacrilamide ratio 37.5:1) was prepared in 65 mM Tris-acetate (pH 6.8) containing 0.1% SDS.
SDS/PAGE and Western Blotting.
Harvested cells were lysed in lysis buffer [20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EGTA, and 1% Triton X-100] containing 1× protease inhibitor mixture (Protease Inhibitor Mixture Tablets; Roche), and the protein concentration of the cell lysate was quantified with a Protein Quantification kit (Dojindo) according to the manufacturer's instructions. Equal amounts of protein (≈20 μg) were mixed with 4× sample buffer (0.25 M Tris-HCl, 40% glycerol, 8% SDS, 0.04% bromophenol blue, and 8% β-mercaptoethanol), boiled at ≈100 °C for 5 min, and then separated by SDS/PAGE with modified polyacrylamide gels. Tris/Tricine/SDS buffer (BioRad) was used as running buffer, and gel electrophoresis was performed at 50 V for 2 h followed by 100 V for ≈30 h at 4 °C. After electrophoresis, separated proteins were electrophoretically blotted onto polyvinylidene fluoride membranes (Immobilon P; Millipore) in transfer buffer [25 mM Tris-HCl (pH 8.3), 192 mM glycine, 10% methanol, and 0.02% SDS] at 150 mA for 60 min. The membranes were blocked for 1 h in blocking solution [5% nonfat milk in PBS-T buffer (PBS containing 0.1% Tween-20)], incubated with 1/3,000 diluted mouse anti-huntingtin protein monoclonal antibody (clone 1HU-4C8) [MAB2166 (Millipore)] at 4 °C overnight, washed with PBS-T buffer, and further incubated with 1/5,000 diluted horseradish peroxidase-conjugated goat anti-mouse IgG (Jackson Immunoresearch Laboratories) for 1 h at room temperature. Antigen–antibody complexes were visualized using Immobilon Western Chemiluminescent HRP Substrate (Millipore). After detection of signals, the membranes were subjected to antibody removal in Re-Blot Plus strong antibody stripping solution (Chemicon) followed by washing in PBS-T buffer, and then incubated with 1/10,000 diluted anti-α-tublin antibody (Sigma-Aldrich). Subsequent processes were the same as described above.
Fluorescent Labeling of Synthetic Oligonucleotides.
Synthetic (CTG)n oligonucleotides (1 μg) each with a different number of CTG repeats were labeled with the Alexa Fluor 488 or 647 fluorescent substrates using the appropriate ULYSIS Nucleic Acid Labeling Kits (Molecular Probes) according to the manufacturer's instructions. Alexa Fluor 488- or 647-labeled (CTG)20 oligonucleotide (100 ng) was mixed with a competitor, 100 ng of a (CTG)n oligonucleotide labeled with the other fluorescent dye. These mixtures were prepared and tested by the pull-down method. The fluorescent intensity of the oligonucleotides recovered by the pull-down method was measured using a NanoDrop-3300 fluorospectrometer (Thermo Scientific).
Construction of Reporter Alleles.
Reporter alleles were constructed as described previously (4). Briefly, phRL-TK (Promega) and pGL3-TK plasmids encoding the Renilla and Photinus luciferase genes, respectively, were digested with XbaI and NotI and subjected to ligation with synthetic oligonucleotide duplexes corresponding to the rs363099, rs362331, rs362273, and rs362272 SNP alleles. The oligonucleotide sequences for the construction of reporter alleles are indicated in Table S7.
Transfection and Reporter Assay.
Transfection and reporter assay were carried out as described previously (4). Briefly, 300 ng of pGL3-TK-backbone plasmid, 50 ng of phRL-TK-backbone plasmid, 100 ng of pSV-β-galactosidase control vector (Promega), and 20 nM (final concentration) of each siRNA duplex were cotransfected into HeLa cells using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. Twenty-four hours after transfection, cell lysate was prepared, and the expression levels of luciferase and β-galactosidase were examined by Dual-Luciferase reporter assay system (Promega) and Beta-Glo assay system (Promega), respectively. The luminescent signals were measured using a TD-20/20 luminometer (Promega).
For examination of dose-dependent inhibition of siRNA (IC50 of siRNA), the day before transfection, HeLa cells were trypsinized, diluted with fresh medium without antibiotics, and seeded into 96-well culture plates at a cell density of 1 × 105 cells/cm2. The pGL3-TK-backbone plasmid (60 ng), phRL-TK-backbone plasmid (10 ng), and pSV-β-galactosidase control plasmid (20 ng) were cotransfected with a defined concentration of a single siRNA [0, 0.001, 0.005, 0.02, 0.08, 0.32, 1.25, 5, 10, or 20 nM (final concentration)] using Lipofectamine 2000 transfection reagent (Invitrogen) and plated into separate wells. Twenty-four hours after transfection, cell lysate was prepared, and the expression levels of luciferase and β-galactosidase were measured with the Dual-Luciferase reporter assay system (Promega) and the Beta-Glo assay system (Promega), respectively, in the Fusion Universal Microplate Analyzer (PerkinElmer). The data were fitted to the Hill equation (Hill coefficient; n = 1), and IC50 values were determined.
Statistical Analysis.
To examine the allele discrimination capability of the pull-down method, the experiments with fluorescent-labeled (CTG)n oligonucleotides (described above) were carried out. The fluorescent intensity of the oligonucleotides was measured before and after the pull-down step. The fluorescent intensity recovered by the pull-down method was normalized to that before pull-down, and ratios of normalized fluorescent intensities of the (CTG)n oligonucleotides as competitors to those of the opposed (CTG)20 oligonucleotide were further normalized to the ratio of the Alexa 488-labeled (CTG)20 vs. Alexa 647-labeled (CTG)20 oligonucleotides. The resultant ratios were first evaluated by one-way ANOVA. When significant differences were indicated by ANOVA, Dunnett's multiple comparison test (P < 0.05) was performed by comparing with the normalized ratio (the relative value = 1) of the Alexa 488-labeled (CTG)20 vs. Alexa 647-labeled (CTG)20 as a standard.
Cloning of the Full-Length ORF Regions of the HTT Alleles.
The cDNAs derived from C0142 and C0221 lymphoblastoid cells were subjected to PCR amplification using a TaKaRa LA Taq polymerase (TaKaRa Bio) and the HD1-F and HD1-ORF-R primers designed previously (16):
HD1-F: 5′-TATAGAATTCGGGAGACCGCCATGGCGAC-3′
HD1-ORF-R: 5′-TCAAGCGGCCGCTCAGCAGGTGGTGACCTTG-3′
The GeneAmp PCR system 9700 (Applied Biosystems) was used as the thermal cycler, and the thermal cycling profiles were as follows: heat denaturation at 95 °C for 1 min, 30 cycles of amplification including denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 12 min. The PCR products were examined by agarose gel electrophoresis followed by ethidium bromide staining (Fig. S2A). The PCR product of ≈9.4 kb was purified from the gels using a PCR & Gel purification kit (BEX) and inserted into the pCR-XL-TOPO plasmid with a TOPO XL PCR cloning kit (Invitrogen) according to the manufacturer's instructions. The resultant plasmid DNA was examined by restriction enzyme digestion with EcoRI and NotI, followed by agarose gel electrophoresis analysis (Fig. S2 B–D). For examination of the CAG trinucleotide repeats and cSNPs, the PCR-based analyses using the HTT exon 1 and exon 1-2 primer sets and the TaqMan SNP Genotyping assays as described above were carried out.
Supplementary Material
Acknowledgments
We thank Drs Y. Nagai, H.A. Popiel, N. Fujikake, and Y. Saitou for their helpful advice and discussion. This work was supported by research grants from the Ministry of Health, Labor and Welfare of Japan and from the National Hospital Organization, by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science, and by a Grant-in-Aid for Research Activity start-up from the Japan Society for the Promotion of Science.
Footnotes
Conflict of interest statement: H.H. and M.T. have a pending patent on the method used in this article.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012153107/-/DCSupplemental.
References
- 1.Bauer PO, Nukina N. The pathogenic mechanisms of polyglutamine diseases and current therapeutic strategies. J Neurochem. 2009;110:1737–1765. doi: 10.1111/j.1471-4159.2009.06302.x. [DOI] [PubMed] [Google Scholar]
- 2.Davidson BL, Paulson HL. Molecular medicine for the brain: Silencing of disease genes with RNA interference. Lancet Neurol. 2004;3:145–149. doi: 10.1016/S1474-4422(04)00678-7. [DOI] [PubMed] [Google Scholar]
- 3.Shao J, Diamond MI. Polyglutamine diseases: Emerging concepts in pathogenesis and therapy. Hum Mol Genet. 2007;16:R115–R123. doi: 10.1093/hmg/ddm213. [DOI] [PubMed] [Google Scholar]
- 4.Ohnishi Y, Tokunaga K, Kaneko K, Hohjoh H. Assessment of allele-specific gene silencing by RNA interference with mutant and wild-type reporter alleles. J RNAi Gene Silencing. 2006;2:154–160. [PMC free article] [PubMed] [Google Scholar]
- 5.Schwarz DS, et al. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hohjoh H. Allele-specific silencing by RNA interference. Methods Mol Biol. 2010;623:67–79. doi: 10.1007/978-1-60761-588-0_4. [DOI] [PubMed] [Google Scholar]
- 7.Huang H, et al. Profiling of mismatch discrimination in RNAi enabled rational design of allele-specific siRNAs. Nucleic Acids Res. 2009;37:7560–7569. doi: 10.1093/nar/gkp835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohnishi Y, Tamura Y, Yoshida M, Tokunaga K, Hohjoh H. Enhancement of allele discrimination by introduction of nucleotide mismatches into siRNA in allele-specific gene silencing by RNAi. PLoS ONE. 2008;3:e2248. doi: 10.1371/journal.pone.0002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W, et al. Linking SNPs to CAG repeat length in Huntington's disease patients. Nat Methods. 2008;5:951–953. doi: 10.1038/nmeth.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Bilsen PH, et al. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington's disease patient-derived fibroblasts. Hum Gene Ther. 2008;19:710–719. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- 11.Snell RG, et al. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat Genet. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 12.Walker FO. Huntington's disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 13.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 14.Lombardi MS, et al. A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Pfister EL, et al. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hohjoh H, et al. Molecular cloning and characterization of the common marmoset huntingtin gene. Gene. 2009;432:60–66. doi: 10.1016/j.gene.2008.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.