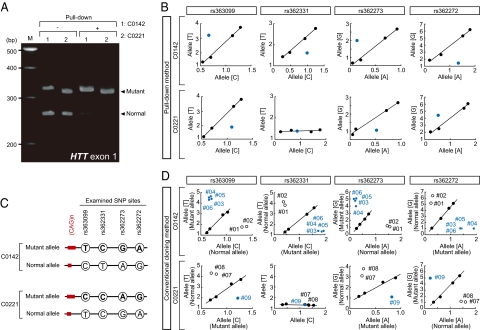
Fig. 2.
Biased recovery of disease-causing HTT alleles. (A) cDNAs prepared from two HD cases (C0142 and C0221) were subjected to the pull-down method (+), and then the length of the CAG trinucleotide repeat array in HTT was investigated by PCR analysis. Untreated cDNAs (−) were also examined as a control. (B) cDNAs (C0142 and C0221), which were the same as in A, were subjected to the TaqMan SNP Genotyping assay, and the results were plotted in blue. Standard curves (indicated in black) were constructed by the same SNP typing using serially diluted (0.01, 1, 10 ng) untreated cDNAs. The examined coding SNP (cSNP) sites are indicated. x- and y-axes indicate relative fluorescent intensity obtained from the TaqMan probes against the indicated alleles, respectively, and allelic nucleotides at the cSNP sites are also shown in parentheses. (C) Linkage between the expanded CAG trinucleotide repeats and cSNPs was estimated in each HD case according to the results of the cSNP typing in B. (D) Confirmatory experiments by conventional methods. The same SNP typing as in B was carried out using the isolated plasmids carrying the HTT alleles, which were obtained by conventional methods (Fig. S2). Dots in blue and open circles represent the results of the plasmids containing the mutant and normal HTT alleles, respectively. Indicated clone (plasmid) numbers correspond to those indicated in Fig. S2.