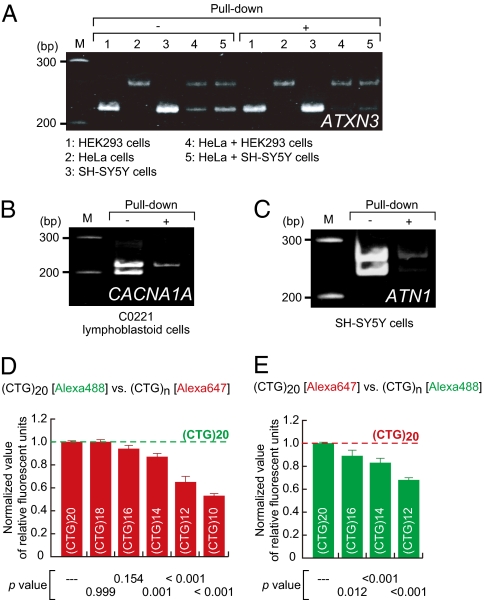
Fig. 3.
Pull-down of (A) ATXN3, (B) CACNA1A, and (C) ATN1 cDNAs carrying longer CAG trinucleotide repeats. cDNAs were prepared from indicated human cell lines and treated by the pull-down method. The recovered cDNAs were examined by PCR analysis and assessed CAG trinucleotide repeat array length in each gene as in Fig. 2A. Because the CAG trinucleotide repeat arrays in ATXN3 (A) were homozygous in all of the samples investigated, mixtures of the HEK293 and HeLa cDNAs (Sample 4) and of the HeLa and SH-SY5Y cDNAs (Sample 5) were prepared and then examined. (D and E) Allele discrimination capability of the pull-down method was examined. The Alexa 488- and Alexa 647-labeled CTG oligonucleotides with various numbers of the CTG trinucleotide repeats, which are complementary to the biotin-CAG RNA probe, were used as targets. The number (n) of the CTG repeats is indicated as (CTG)n. (CTG)20 oligonucleotides, the longest CTG repeats arrays (20 repeats), were labeled with Alexa 488 (D) or Alexa 647 (E) and individually mixed with a competitor, a (CTG)n oligonucleotide labeled with the other fluorescent dye; these mixtures were then subjected to the pull-down assay. The fluorescence intensities of the oligonucleotides recovered by the pull-down method were examined. Ratios of normalized fluorescent intensities of the competitors to that of the (CTG)20 oligonucleotide are shown. Data are averages of six independent tests. Error bars indicate SD. P values were calculated by Dunnett's multiple comparison test.