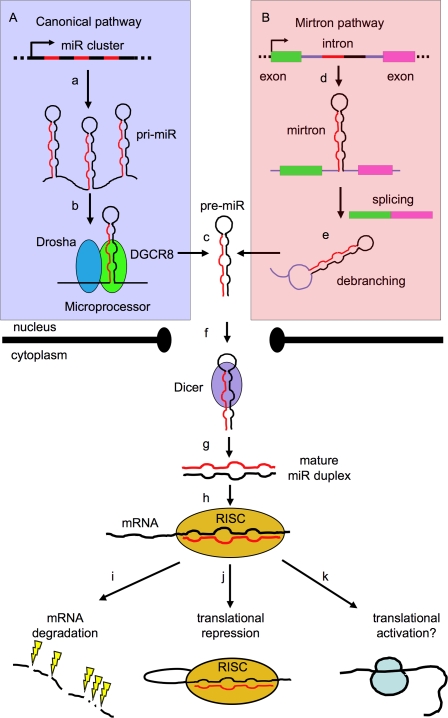
Figure 1.
Biogenesis of miRs. (A) The canonical pathway is shown boxed in violet. Most miR promoters characterized to date are typical of RNA polymerase II transcribed genes (Lee and Dutta 2009), and give rise to a 5′-capped, spliced, and polyadenylated primary precursor (pri-miR) (a), formed by one or several concatenated hairpin structures (Lee et al. 2002, 2004; Altuvia et al. 2005) consisting of a stem (containing a miR) and a terminal loop. The pri-miR is recognized cotranscriptionally in the nucleus by the Microprocessor complex (b), whose catalytic core (formed by the Drosha and DGCR8 proteins) processes the base of the stem of the pri-miR and trims away the flanking sequences to release an intermediate stem–loop structure (known as pre-miR) of ∼70 nt (c). (B) A second, noncanonical pathway is shown boxed in pink. (d) Some animal miRs (“mirtrons”) are embedded in mRNA introns and completely bypass processing by Microprocessor. (e) Instead, they are processed by splicing and debranching (Okamura et al. 2007). In both cases, the resulting pre-miR is transported to the cytoplasm (via the exportin–RanGTP pathway) (f) (Yi et al. 2003), where the loop is further processed by the Dicer complex to release a mature miR duplex (g), which is finally incorporated into the RISC (h) (Hammond et al. 2000). The RISC recognizes the miR duplex, unwinds it, selects the miR strand while degrading the passenger strand, and mediates recognition of the target mRNA. The downstream regulatory effect is degradation of the mRNA (the prevalent mechanism in mammalian cells (i) (Guo et al. 2010) or its translational repression (j) (Mourelatos et al. 2002; Pratt and MacRae 2009), although instances of up-regulation have also been reported (k) (Vasudevan et al. 2007).