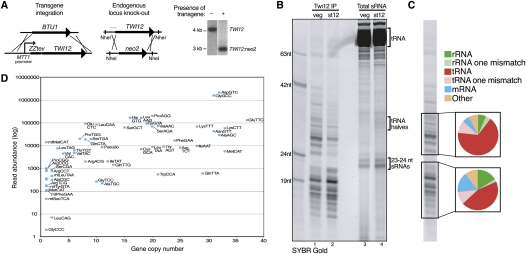
Figure 1.
Twi12 association with tRNA fragments. (A) The schematics show the strategy of ZZTwi12 transgene knock-in and endogenous TWI12 locus knockout; the latter is evidenced with Southern blot data using NheI-digested genomic DNA at right. The position of the Southern blot probe is indicated in the TWI12 locus schematic with a thin gray line. (B) RNAs copurified with Twi12 in vegetative growth (veg) or after 12 h of starvation (st12) were stained directly with SYBR Gold in comparison with size-selected total RNA from the same cultures. (C) The composition of sRNA library sequencing reads is depicted for reads with either a perfect match to the genome or a single internal mismatch as indicated. (D) Read numbers for tRNA fragments copurified with Twi12 were plotted relative to tRNA gene copy number as an approximation of full-length tRNA abundance. The labels of some data points at left were removed for clarity: mtGluTCC, SecTCA, ThrCGT, mtHisGTG. Pseudo indicates combined reads from the 11 annotated tRNA pseudogenes, and mt indicates mitochondrial.