Figure 2. Canonical model of mode of action of core Hsp70 machinery in protein folding and Hsp70 structure.
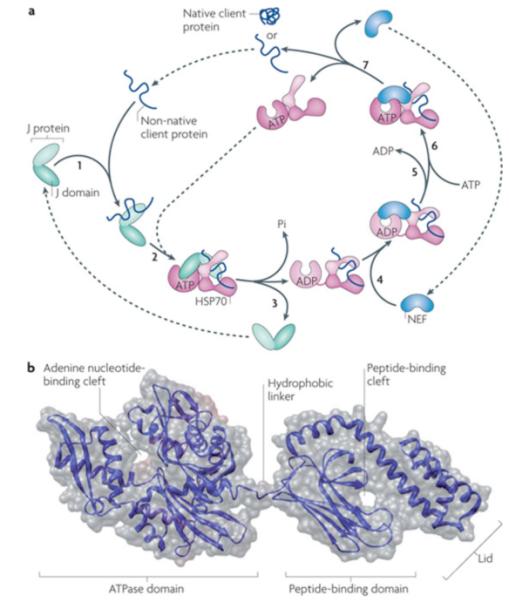
a|Canonical mode of functioning of the Hsp70 core machine based on in vitro refolding studies of denatured proteins. [1] J-protein binds to client protein via its peptide-binding domain and [2] interacts with Hsp70(ATP) via its J-domain (J). [3] The client rapidly, but transiently, interacts with the “open” peptide binding site of Hsp70. ATP hydrolysis is stimulated by both the J-domain and client causing a conformational change in Hsp70 closing the helical lid over the cleft, stabilizing client interaction. J-protein leaves the complex. [4] Nucleotide exchange factor (NEF), which has a higher affinity for Hsp70(ADP) than Hsp70(ATP), binds Hsp70; [5] ADP dissociates through distortion of the ATP binding domain, after which [6] ATP binds to Hsp70. [7] Client is released because of its low affinity for Hsp70(ATP). ATP-binding to Hsp70 is favored since cellular ATP concentrations are typically much higher than those of ADP. If the native state of the client is not attained upon release, J-protein rebinds to exposed hydrophobic regions and the cycle begins again. Typically such reiterative binding is required. b| Hsp70 structure with ADP bound to the nucleotide binding domain135 (PDB code #2KHO). The ATPase domain and peptide-binding domain are connected via a short flexible linker. These domains dock when in the ATP-bound state, which is also thought to displace the lid, allowing easy access and egress of the client protein from the cleft 17, 136.
