Figure 3. Diversity in domain architecture of J-proteins from the yeast S. cerevisiae and H. sapiens.
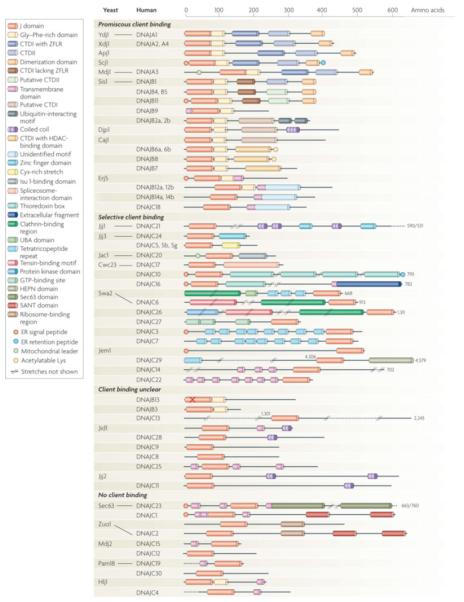
Members are clustered according to known or presumed client binding ability. Functional orthologs are connected by lines. For clarity, not all known domains are indicated and some differences between yeast and human orthologs are not indicated. For more detailed information see Supplementary Tables I, II, where information about the presumed localization and function of all members is also provided. Regions of Class I and II J-proteins: G/F regions, of which the functional relevance is disputed (see main text) are defined as segments containing more than 5 glycines or/and phenylanines in the first 25 amino acids C-terminal to J-domain. C-terminal domain 1 (CTD1) - canonical type I members have Zinc finger like region (ZFLR); type II members lack a ZFLR. However, type II often have cysteine rich stretches and/or binding site for Histone Deacetylases (HADCs). The indicated dimerization domain (DD) has been firmly established only for a few type I and II (Ydj1, DNAJA1, Scj1, Sis1); for the others this domain is presumed for simplicity. “X” in DNAJB13 indicates the lack of the canonical HPD motif in the J-domain. For reasons of space, some members are not drawn at full size (indicated by numbers); //..// indicate stretches not shown.
