Figure 4. J-domain and client protein binding domain structures.
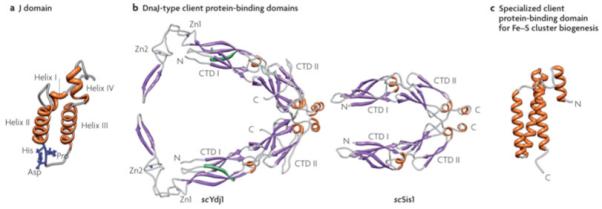
a| J-domains contain four alpha-helices, with the central ones forming a coiled-coil motif around a hydrophobic core137. The invariant HPD tripeptide located in the loop between helices II and III is critical for ATPase stimulation and in vivo function12. Residues in helix II and within the neighboring loop, including the HPD, form an Hsp70 interaction face. b| Class I Ydj1 and Class II Sis1 have similar client protein binding domains, the “DnaJ-type”. The structure of amino acids 102-350 of Ydj126, 29 and amino acids 180-343 of Sis130 are shown. Both have J-domains at their N-termini, followed by a glycine/phenyalanine-rich (G/F) region. No full-length structure of a Class I or Class II J-protein has been obtained, presumably because of the flexibility of the G/F regions. Both Ydj1 and Sis1 are dimers, with the dimerization domain at their C-termini (C). Each monomer of Ydj1 and Sis1 has two adjacent domains that are similar structure, being predominantly composed of beta-sheets. These are often referred to as “C-terminal domain” (CTD1) and CTD2. Ydj1 (like DnaJ) has two zinc fingers (Zn1 and Zn2), which extend out from CTD1. In addition, Ydj1 has a CAAX motif for farnesylation, a modification important for membrane localization and binding of some client proteins138 139. c|Jac1 (called HscB in E. coli), which is critical for Fe-S cluster biogenesis, has a specialized client protein binding domain that has neither sequence nor structural similarities to a DnaJ-type140; amino acids 63-171 of HscB are shown. The face pointing outwards interacts with the Fe-S cluster scaffold Isu141.
