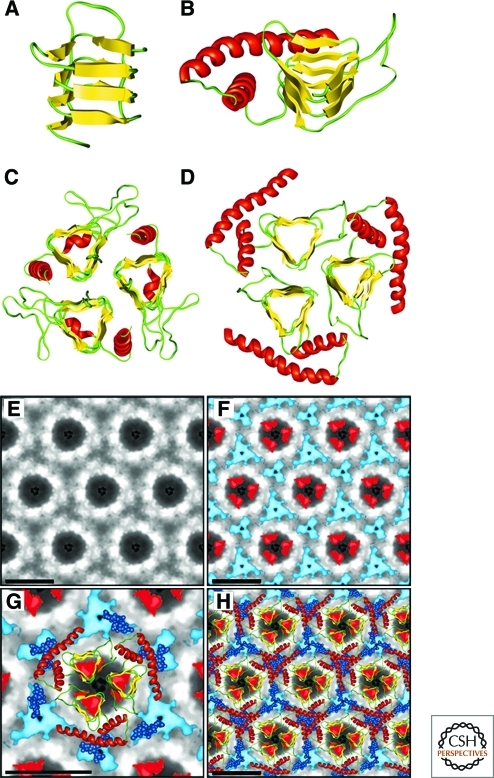
Figure 5.
Structural models of PrPSc. (A) Residues 89–174 of PrP threaded into a left-handed β-helix based on UDP N-acetylglucosamine O-acyltransferase from Escherichia coli (PDB ID code 1LXA). (B) Model of the monomer of PrP 27–30 with the α-helical region (residues 177–227) as determined by NMR spectroscopy shown in red. (C) The crystal structure of the trimeric carbonic anhydrase from Methanosarcina thermophila. (D) Trimeric model of PrP 27–30 built by superimposing three monomeric models onto the structure shown in C. (E) Projection map of PrP 27–30 obtained by processing and averaging three independent 2D crystals of PrP 27–30. (F) Statistically significant differences between PrP 27–30 and PrPSc106 overlaid onto the projection map of PrP 27–30. The differences attributed to the internal deletion of PrPSc106 (residues 141–176) are shown in red; the differences in glycosylation between PrP 27–30 and PrPSc106 are shown in blue. (G) Superimposition of the trimeric left-handed model onto the EM maps. The trimeric left-handed α-helical model of PrP 27–30 is superimposed on a 1:1 scale with the electron crystallographic maps of PrP 27–30. (H) The scaled trimeric model was copied onto the neighboring units of the crystals to show the crystallographic packing suggested by the model. Bars in panels E–H represent 50 Å. Reprinted with permission, from Govaerts et al. 2004a.