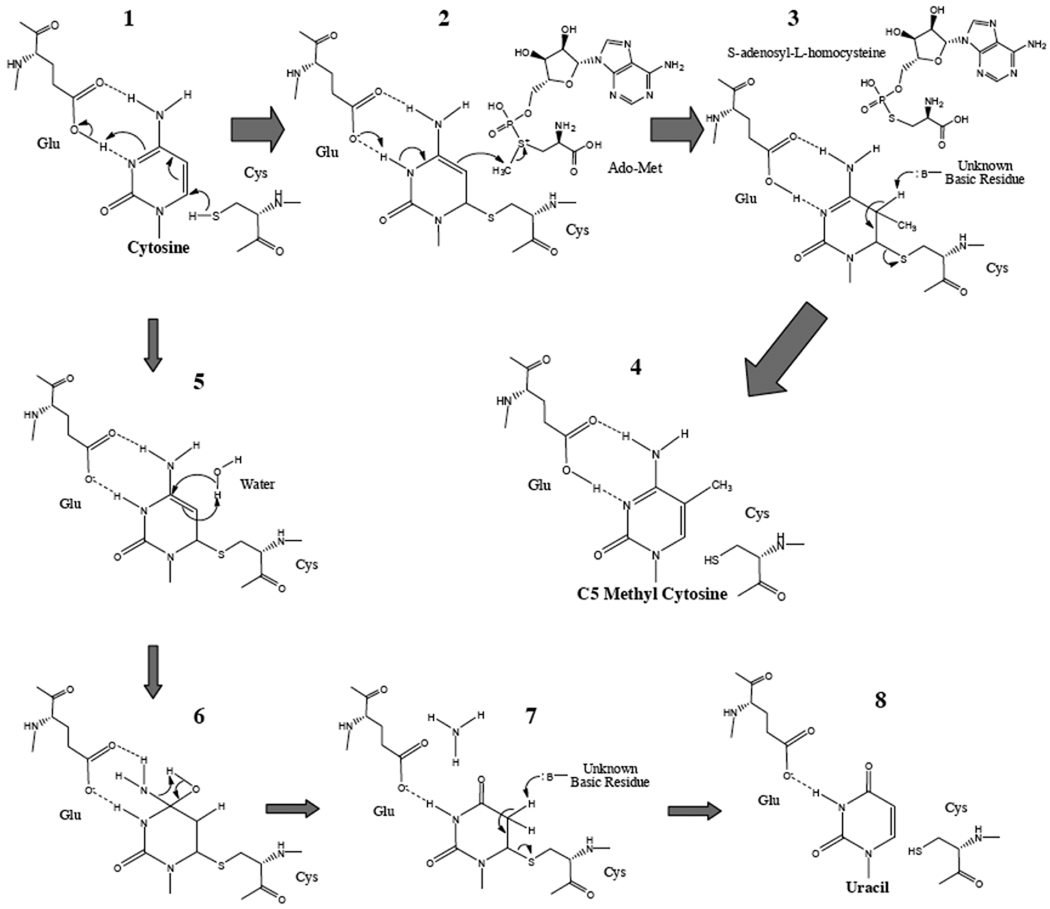
Fig. 2.
The chemical mechanism of cytosine C5 methylation (1–4) and its possible side-effect reactions (5–8). The sulfhydryl group at the side chain of the key cysteine residue, in the active-site loop of DNA MTase, could nucleophilically attack to the C6 position of cytosine followed by transient protonation of the N3 nitrogen atom via the glutamate residue in conserved motif VI of DNA MTase (1–2). Consequently, aromaticity of cytosine is broken so as to strengthen activity of nucleophilic attack of the C5 position in cytosine to the methyl group in the sulfonium centre of Ado-Met (2). Following the methyl group transferred to the C5 position of cytosine, the N3 position in cytosine is re-deprotonated (b). To release enzyme moiety covalently bound in the C6 position of cytosine, the C5 position could be deprotonated with assistance of unknown residues in the enzyme (3). Following the β-elimination reaction involved in C5 and C6 positions in cytosine, C5-methyl cytosine could finally be formed (4). Under certain conditions (e.g. Ado-Met inadequate), water can penetrate into the active site of DNA MTase towards the relatively localized π electron in the activated cytosine (5). Then the C4 position in cytosine is hydroxylated so that deamination reaction is triggered (6). The amino group in the C4 position of cytosine then is replaced to form a carbonyl oxygen atom (7). The conversion of cytosine to uracil is accomplished (7–8).