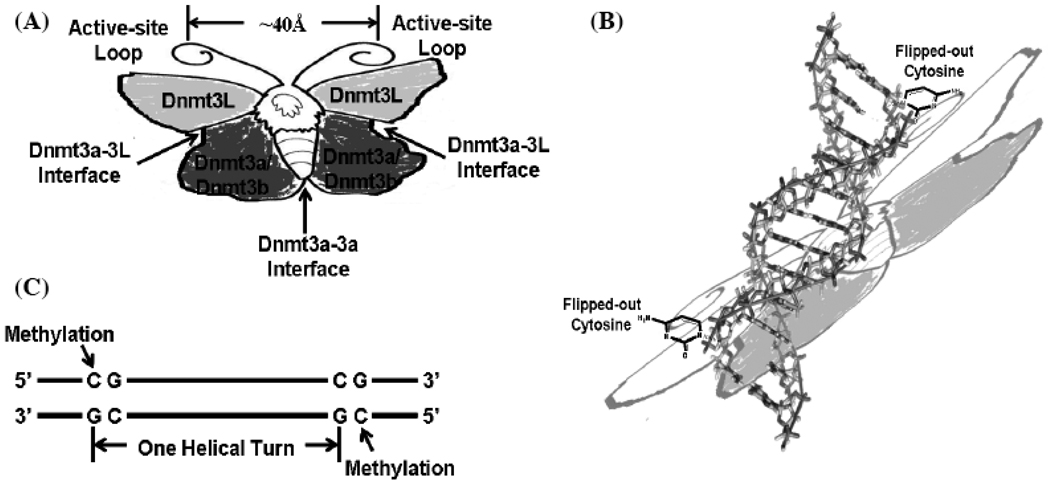
Fig. 4.
CG spacing pattern in Dnmt3-mediated de novo DNA methylation. (A): Two Dnmt3a/3b and two Dnmt3L can form a tetramer complex, in which there may be three interfaces: one central Dnmt3a-Dnmt3a homodimer interface and two lateral Dnmt3a-Dnmt3L heterodimer interfaces. The overall Dnmt3L-(Dnmt3a)2-Dnmt3L complex can be proposed as an elongated, butterfly-like structure, in which the distance between two active-site loops in each Dnmt3a/3b is approximately 40 Å apart. (B), (C): Dnmt3L-(Dnmt3a)2-Dnmt3L complex can target the unmethylated DNA duplex to simultaneously de novo methylate the two flipped-out cytosines in CG sites of both the strands that is one-helical-turn apart.