Figure 1.
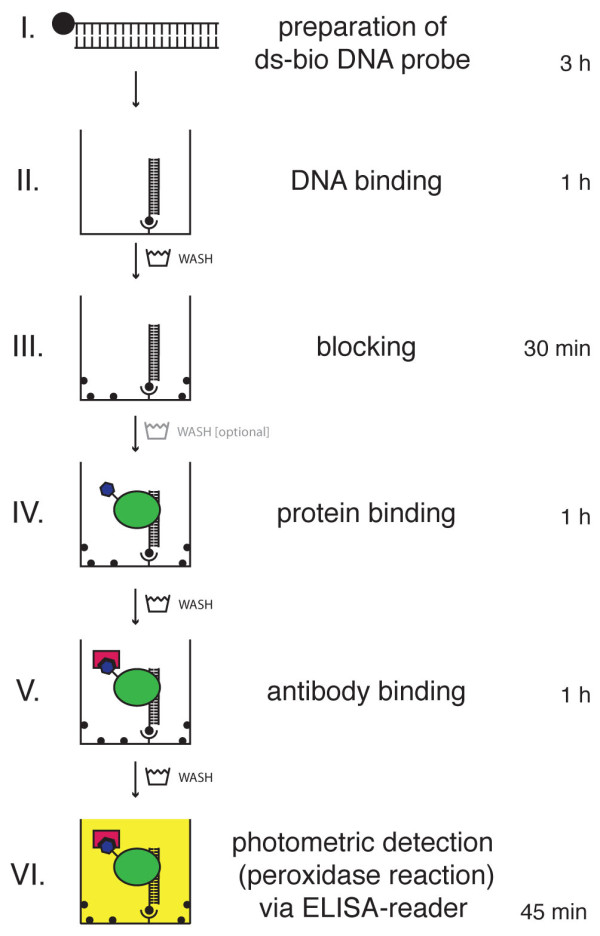
Schematic workflow of the DPI-ELISA. Double stranded biotinylated (ds-bio) DNA-probes (I) are immobilised on a streptavidin-coated microtiter plate (II). After blocking the plate with an appropriate reagent (III), incubation with a crude protein extract from E. coli containing the epitope tagged protein under investigation is performed (IV). The epitope tagged protein is retained inside the well by physical binding to the immobilized DNA and, thus, can be detected with appropriate antibodies - which are conjugated with horseradish peroxidase in our experiments (V). Finally, peroxidase substrate (OPD) is added for the colorimetric quantification of specifically bound transcription factors to dsDNA-probes (VI). Between each of the workflow steps, at least three washing steps of the microtiter plate are performed; washing between steps III and IV is optional. Incubation times and approximate duration of the photometric detection step (peroxidase reaction) are given at the right hand side.
