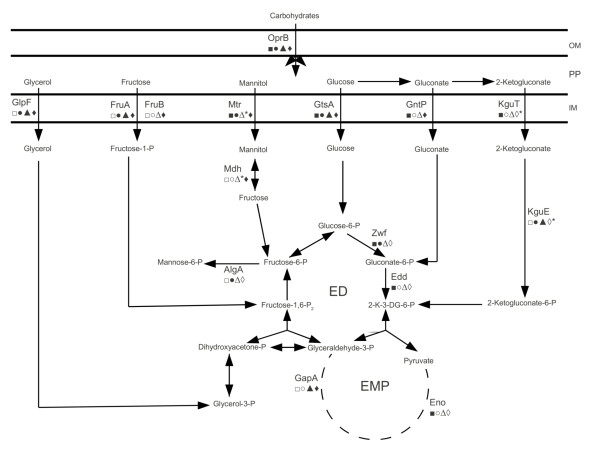
Figure 2.
Predicted Crc regulon of carbohydrate metabolism in Pseudomonas. Selected genes involved in carbohydrate transport and metabolism are shown along with their status vis a vis (predicted) Crc regulation. Genes from P. aeruginosa (squares), P. fluorescens (circles), P. putida (triangles) and P. syringae (diamonds) are shown, with filled/unfilled symbols indicating that the target in that species is/is not predicted to be regulated by Crc. An asterisk (*) after a symbol indicates where an orthologous locus is absent in the relevant species. OM - outer membrane; PP - periplasm; IM - inner membrane; ED - Entner-Doudoroff pathway; EMP - Embden-Meyerhoff pathway; 2-K-3-DG-6-P - 2-keto-3-deoxygluconate-6-phosphate. OprB - carbohydrate porin B; GlpF - glycerol transporter; FruAB - fructose phosphotransferase system; Mtr - mannitol transporter subunit; GtsA - glucose transporter subunit; GntP - gluconate transporter; KguT - 2-ketogluconate transporter; Mdh - mannitol dehydrogenase; AlgA - mannose-6-P isomerase; Zwf - glucose-6-P dehydrogenase; Edd - gluconate-6-P dehydratase; KguE - xylose isomerase; GapA - glyceraldehyde-3-P dehydrogenase; Eno - phosphopyruvate hydratase. Some steps of the Embden-Meyerhoff pathway are abbreviated with a dashed line for clarity.