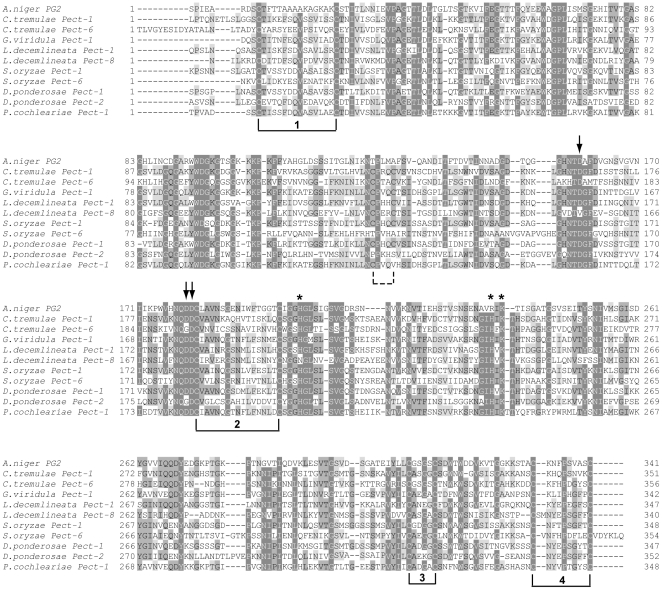
Figure 3. Predicted amino acid alignments of selected GH28 beetle enzymes.
The amino acid sequence of the endopolygalacturonase II from Aspergillus niger (for which the crystal structure has been resolved) is used as a reference sequence. The catalytic residues, predicted from the A. niger sequence, are marked with arrows. Asp180 and Asp202 (numbering according to the A. niger sequence) act as the catalytic nucleophile/base, and Asp201 is the catalytic proton donor. The amino acid residue corresponding to Asp180 in L. decemlineata Pect-8 is a Val residue. The amino acid residue corresponding to Asp201 in C. tremulae Pect-6 and D. ponderosae Pect-2 is a Gly residue. Such changes may affect the catalytic abilities of the given enzymes. The four conserved disulfide bridges are numbered. An extra two Cys residues found in some sequences can form an extra disulfide bridge indicated by the dashed line.