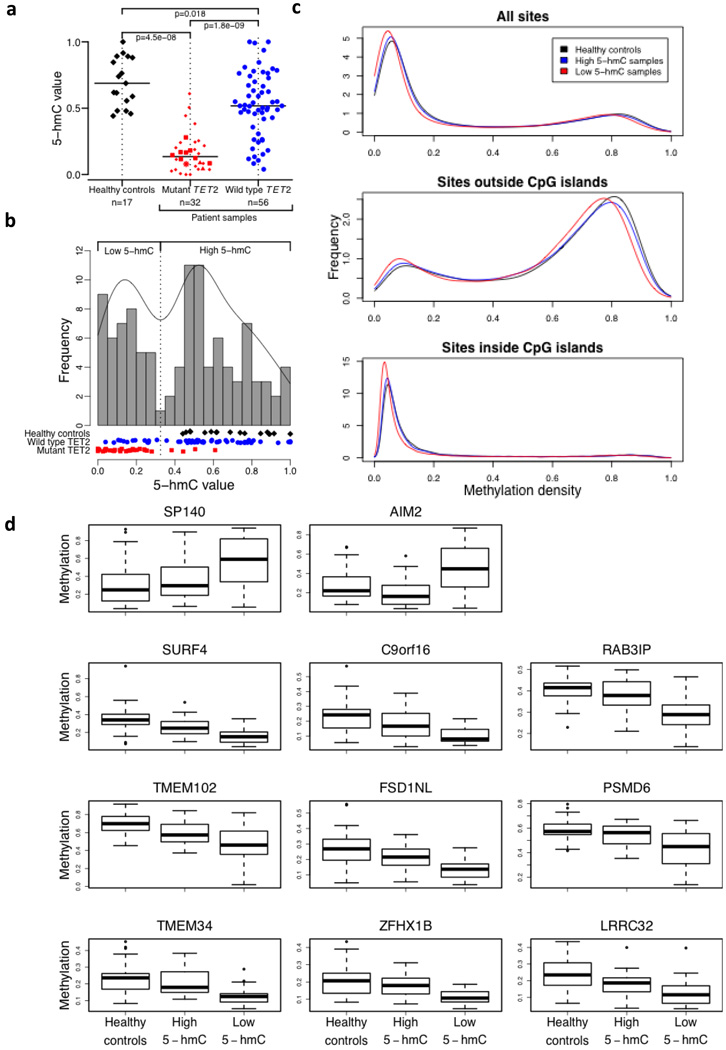
Figure 4. Relation of 5-hmC levels to DNA methylation status.
a, Normalised 5-hmC (CMS) levels in DNA from three different groups: healthy controls (black diamonds), patients with mutant TET2 (red symbols) and patients with wild type TET2 (blue circles). Among TET2 mutants, we distinguish homozygous (squares), hemizygous (triangles), heterozygous (small diamonds) and biallelic heterozygous (star) mutations (for definitions see Online Methods). The horizontal bar indicates the median for each group. The number of samples in each group is indicated.
b, Histogram of normalised 5-hmC (CMS) levels in DNA from healthy donors (black diamonds), patients with mutant TET2 (red rectangles) and patients with wild type TET2 (blue circles). The frequency was calculated based on a Gaussian kernel estimator. The local minimum between both modes was used as a threshold (vertical dotted line) between low and high 5-hmC values.
c, Density of methylation values for healthy controls (black), high 5-hmC samples (blue) and low 5-hmC samples (red) of all sites (top panel), sites outside CpG islands (middle panel) and sites inside CpG islands (lower panel).
d, Boxplot for group-specific methylation for the only two hypermethylated sites (SP140, AIM2; top panel) and the top nine hypomethylated sites (lower panels) between healthy controls and low 5-hmC samples (total number of differentially sites was 2512).