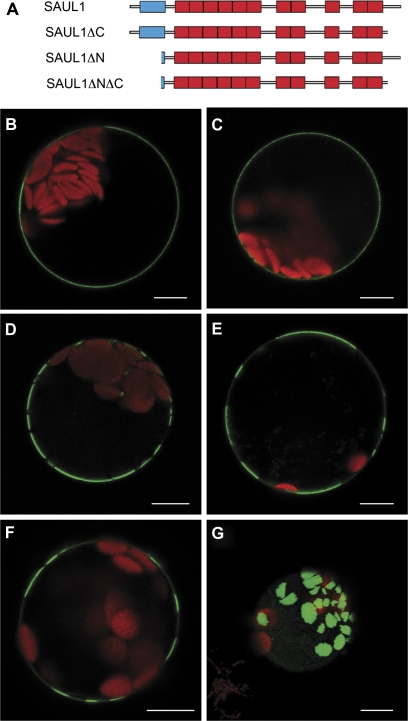
Fig. 3.
Analysis of the contribution of the C- and N-terminus for localization of SAUL1 at the plasma membrane. (A) Schematic representation of SAUL1 deletion constructs. The U-box and ARM repeats are shown in blue and red, respectively. (B) Localization of SAUL1-GFP-HDEL in the plasma membrane indicated that SAUL1 does not require the ER pathway to reach the membrane. (C–E) Localization of SAUL1ΔC–GFP (C), SAUL1ΔNΔC–GFP (D), and SAUL1ΔN–GFP (E) at the plasma membrane. Note the distribution of N-terminally deleted fusion proteins in membrane patches. (F and G) Patchy distribution of GFP–SAUL1. In 25% of transformed protoplasts, GFP–SAUL1 was not equally distributed in the plasma membrane but instead was found in patches. (G) Top view of an Arabidopsis protoplast transformed with the CaMV35S:GFP-SAUL1 DNA construct. The GFP–SAUL1 fusion protein localized to large membrane patches. Autofluorescence of chlorophyll is shown in red. Scale bars represent 10 μm.