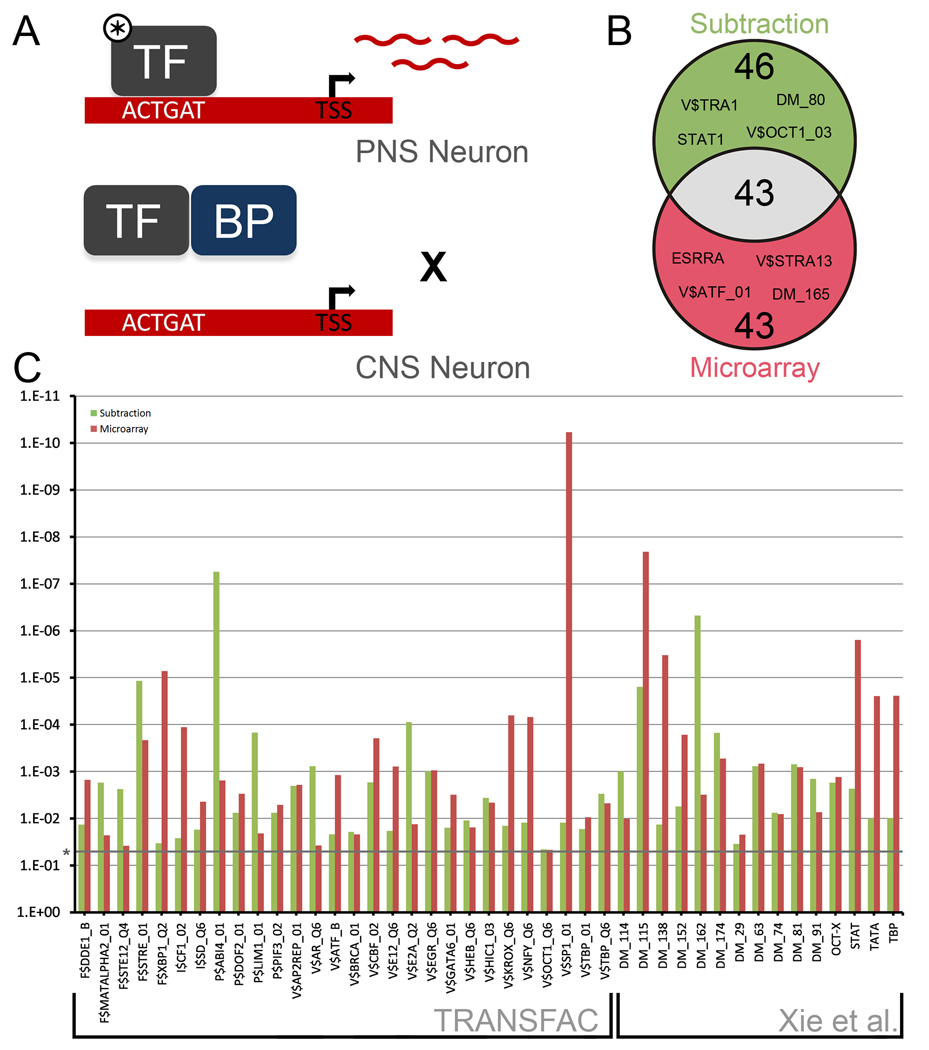
Figure 3. Overrepresented mammalian transcription factor binding sites in the promoters of DRG enriched genes.
(a) Model demonstrating how differentially active TFs may be detected by the analysis of DRG enriched gene promoters. In PNS neurons, an active TF (marked by *) is able to bind target sequences, inducing the transcription of mRNAs that can be detected by the Subtraction and Microarray. In CNS neurons, a TF may be unable to drive expression of target sequences due to a lack of upstream signals or co-activators, the presence of binding proteins dampening activity (BP), or an altered genomic landscape. (b) Venn Diagram of 132 transcription factor binding consensuses found to be significantly overrepresented in the promoter regions of DRG enriched genes. 1200 bp promoter regions (1000bp upstream and 200 bp downstream of TSS) were scanned for consensus binding sites from TRANSFAC 10.2 and from a paper by Xie et al. (2005). (c) 28 TF binding consensuses from TRANSFAC and 15 from Xie et al. (2005) were found to be significantly overrepresented in both the Subtraction and the Microarray data sets. Significance was determined by a two-tailed Fisher’s Exact Test, followed by a Benjamini-Hochberg correction for multiple comparisons.