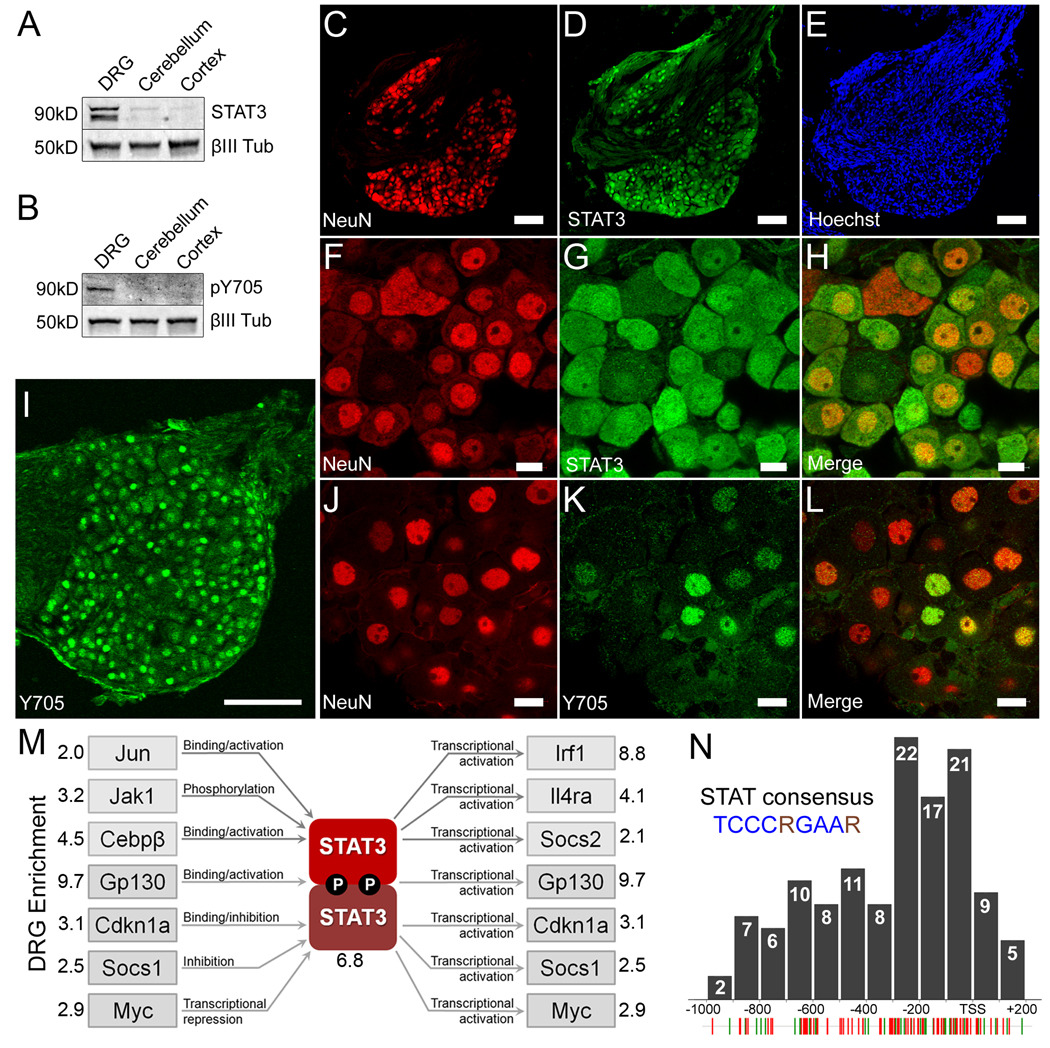
Figure 4. STAT3 is constitutively enriched and active in PNS neurons but not CNS neurons.
(a–b) Representative Western blot of lysates from P7-10 mouse dorsal root ganglia, cerebellum, and cerebral cortex probed for (a) total STAT3 and (b) Y705 phosphorylated STAT3. Neuronal specific βIII-tubulin was used as a loading control. (c–e) STAT3 co-localizes with NeuN but not Hoechst positive Schwann cells in the nerve root. (c) NeuN, (d) total STAT3, (e) the nuclear dye Hoechst. (f–h) High magnification confocal micrograph of (f) NeuN, (g) total STAT3, and the (h) merged image showing co-localization. (i) DRG neurons express nuclear Y705 phosphorylated STAT3 constitutively. (j–l) High resolution confocal micrographs showing (j) Islet 1 staining neuronal nuclei, (k) Y705 STAT3 and (l) the merged image. Scale bars: (c,d,e,i), 100µm; (f,g,h,j,k,l), 10µm. (m) The relative expression of 84 JAK-STAT pathway members in P7-10 dorsal root ganglia and cerebellum was probed using real-time PCR. Multiple factors that regulate and are regulated by STAT3 activity were found to be enriched in DRGs, suggesting the presence of at least one PNS specific STAT3 signaling pathway. (n) Distribution of the STAT consensus binding motif (TCCCRGAAR) within the promoter regions of 126 DRG enriched genes. Individual STAT consensus matches for genes identified by the subtraction (green hatches) and microarray (red hatches) are shown below. Many sites cluster within 200bp upstream of the transcription start site (TSS).