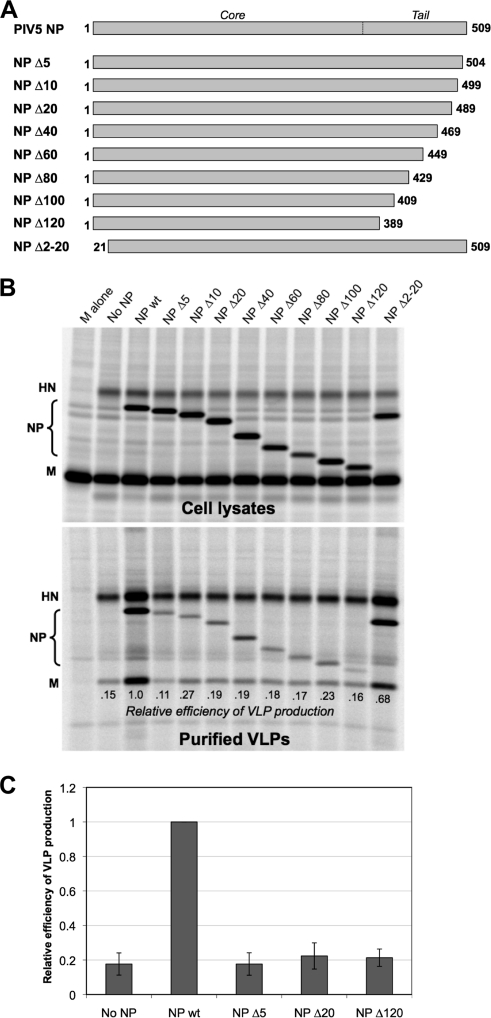
FIG. 1.
Minor truncations to the C-terminal end of PIV5 NP protein disrupt VLP production function. (A) Schematic illustration of truncated PIV5 NP proteins. Numbers indicate the span of amino acid residues that are present in each construct. An approximate boundary between the core and tail regions of NP protein, based on homology with other paramyxovirus nucleocapsid proteins, is indicated. (B) 293T cells were transfected to produce PIV5 M and HN proteins together with the indicated PIV5 NP protein variants. Viral proteins from cell lysates were collected by immunoprecipitation after metabolic labeling of cells. VLPs from culture supernatants were purified by centrifugation through sucrose cushions followed by flotation on sucrose gradients. Viral proteins were separated by SDS-PAGE and visualized using a phosphorimager. Efficiency of VLP production was calculated as the amount of M protein detected in VLPs divided by the amount of M protein detected in the corresponding cell lysate fraction, normalized to the value obtained with wt NP protein. (C) Three independent experiments were performed as described for panel B, and VLP production efficiencies were calculated, with error bars indicating standard deviations.