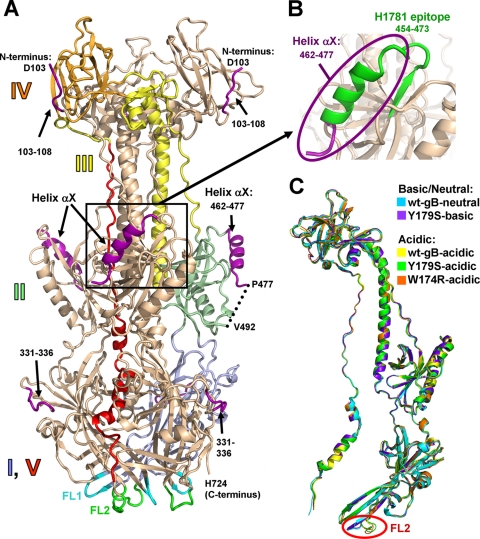
FIG. 1.
(A) W174R-acidic structure (chain A). A single protomer is colored by domain with domain I, pale blue; domain II, pale green; domain III, yellow; domain IV, orange; and domain V, red. Domains are labeled. New regions, unresolved in the previously determined wt-gB-neutral structure (PDB 2GUM), are shown in purple. Fusion loops FL1, residues 175 to 180, and FL2, residues 258 to 264, are shown in cyan and green, respectively. (B) The region of gB containing the now-visible epitope of the neutralizing MAb H1781, residues 454 to 473 (5), is shown in green. The view in panel B is shown in the same orientation as in panel A. (C) Superposition of the four structures determined here—Y179S-basic (chain C), W174R-acidic (chain B), Y179S-acidic (chain A), and wt-gB-acidic (chain A)—onto the previously determined wt-gB-neutral structure (chain A). The same chains are shown in all subsequent structure figures. Single protomers are shown for clarity. All superpositions were performed with Coot (12) using the entire wt-gB-neutral structure as reference molecule. For any pairwise superposition, the root mean square deviation (RMSD) over all Cα atoms is less than 1.25 Å (see Table S2 in the supplemental material). Relative to the colored protomer in panel A, the protomers in panel C are rotated 40° to the viewer's left around the vertical axis.