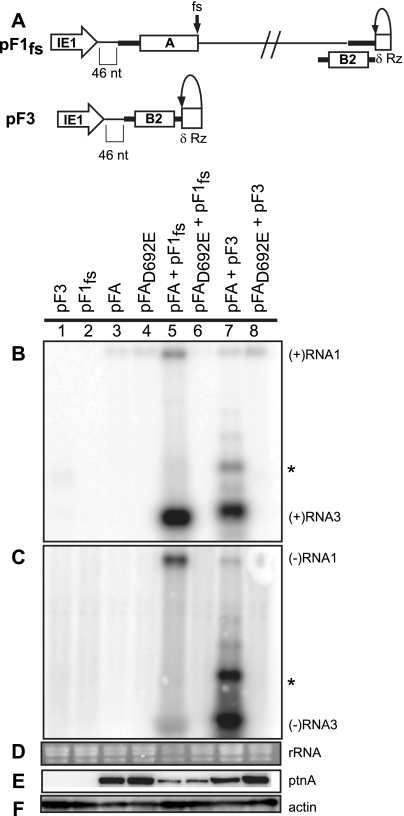
FIG. 5.
trans replication of FHV RNA templates. (A) Schematic of plasmid-directed RNA1fs and RNA3 expression in Drosophila cells. pF1fs is the same as pF1, except that a 4-nt sequence causes a frameshift (fs) that prevents full-length protein A production. pF3 expresses RNA3 from the IE1 promoter with a δRz-mediated authentic 3′ end (δRz). The 5′ ends of pF1fs and pF3 have 46 nonviral nt between the transcription start site and the first viral nucleotide, as was shown for pF1 and pF2. Drosophila cells were transfected with pF3, pF1fs, pFA, pFAD692E, pFA plus pF1fs, pFAD692E plus pF1fs, pFA plus pF3, or pFAD692E plus pF3. (B to D) Total RNA was analyzed by Northern blotting with 32P-labeled cRNA probes for positive-strand RNA1 and RNA3 (B) or negative-strand RNA1 and RNA3 (C). Note that in panel B generally low but somewhat varying levels of the expected pFA- or pFAD692E-generated, nonreplicatable protein A mRNA transcripts are visible in lanes 3 to 8 at a position slightly above the replicated positive-strand RNA1 band in lane 5. As noted in an earlier study (59), the similarly sized, weak band near RNA1 in panel C, lane 7, appears to represent detection of an incompletely denatured hybrid between negative-strand RNA3 and the positive-strand protein A mRNA. This band does not represent negative-strand RNA1 since, despite its high position in the gel, the band is only detected with a strand-specific probe against the common sequences of negative-strand RNA3 and RNA1, but not with a strand-specific probe against the unique 3′-proximal portion of negative-strand RNA1, outside of the sequences shared with RNA3 (59). (D) EtBr-stained rRNA is shown as a loading control. Total protein was analyzed by Western blotting with antibodies against protein A (E) or actin (F). The asterisk represents a slower-migrating form of RNA3 that has been previously observed.