Figure 2.
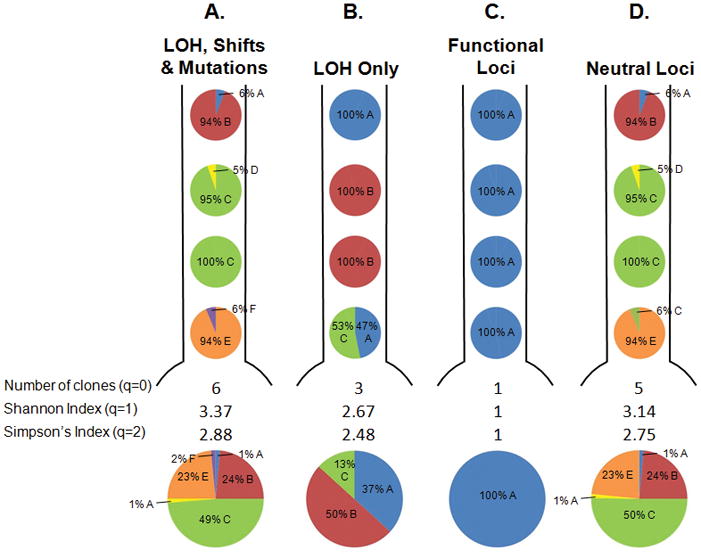
Map of the clones in the BE segment from a single participant from our cohort under different definitions of a clone. The pie charts below each segment represent total % of each clone in the BE segment. Defining clones in different ways alters the distribution and number of clones and the diversity indices. A. Both LOH and changes in microsatellite lengths (shifts) in all 18 microsatellites, as well as sequence mutations in CDKN2A (p16/INK4A) and TP53 (p53), and DNA content are used to define a clone. B. Only LOH lesions in the 18 microsatellites and DNA content are used to define a clone, while shifts and mutations are ignored. C. Only lesions that increase the fitness of a clone are used to define a clone. These lesions include LOH or sequence mutations in CDKN2A or TP53, or hypermethylation of the CDKN2A promoter (20). D. DNA content and lesions that have no detectable effect on the fitness of a clone were used, including shifts in any of the 19 microsatellites and LOH on the q arms of chromosomes 9 and 17 (20). This illustrative patient was chosen because the different definitions of clones lead to different diversity measures, though this was often not the case for other participants in the cohort.
