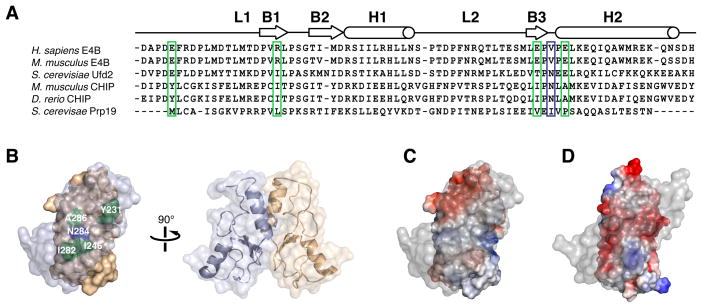
Figure 3. Electrostatic Surface Potentials of E4B and CHIP U-box Domains Affect Oligomerization States.
(A) Sequence alignment of U-box domains from different E3 ubiquitin ligases indicating locations of secondary structure elements. CHIP and Prp19 are dimers while Ufd2 and E4B are monomers. Key CHIP and Prp19 residues mediating homodimerization are highlighted. Hydrophobic residues in CHIP and Prp19 (green box) correspond to charged residues in E4B while an asparagine residue (Asn284) in mouse CHIP (blue box) is replaced by a valine residue (Val1283) in human E4B.
(B) Molecular representation of mouse CHIP homodimer showing the important dimerization interface residues described in A.
(C) Electrostatic surface potential of mouse CHIP U-box homodimer (PDB entry 2C2V) showing predominantly non-polar character at the dimerization interface. Foremost protomer is rendered transparent for clarity.
(D) Electrostatic surface potential of a hypothetical homodimer of E4B U-box showing a negatively charged dimer interface.