Figure 2.
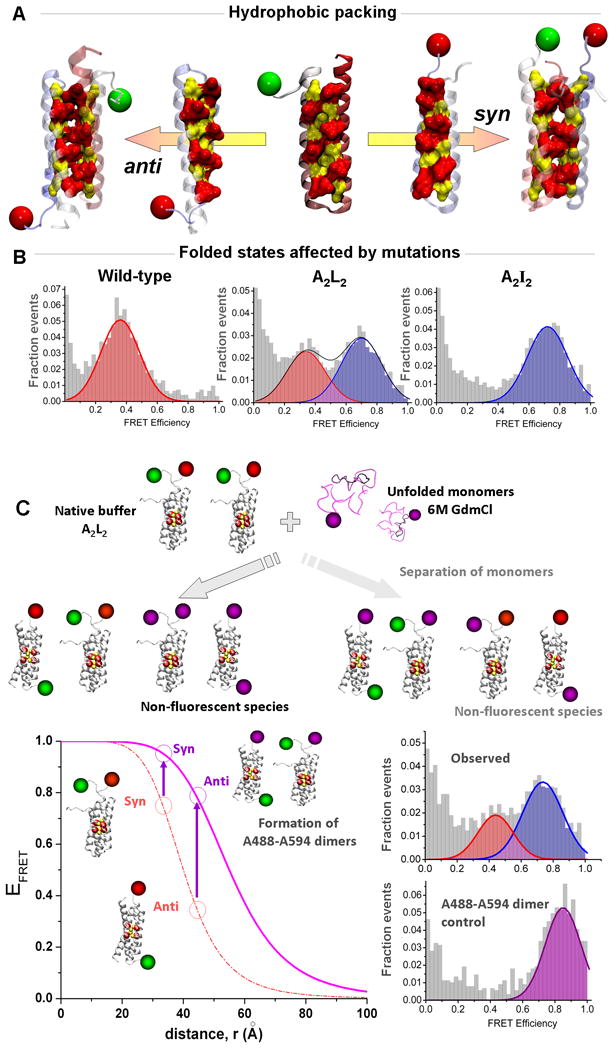
Multicolor smFRET study of the Rop mutants (adapted from reference 51: Gambin et al., Proc. Natl. Acad. Sci., USA, 2009, 106: 10153–10158)
(A) The Repressor of Primers (Rop) complexes are formed by association of two helix-turn-helix motifs. The active enzyme requires that the two monomers bind in an anti conformation to form the proper RNA interface. (B) The symmetry created by multiple mutations within the hydrophobic core of the protein pushes the two monomers to re-arrange into a stable syn conformation, as observed for the A2I2 mutant. Surprisingly, conditions were found where the A2L2 mutant can form coexisting anti and syn, as evidenced by the presence of two FRET peaks. (C) Multi-color smFRET was used to check whether monomers are separating during the interconversion between the two opposite folded states. In a dual-channel format, the experiments took advantage of the difference between two possible dye pairs to detect in a simple manner the formation of mixed species in a competition assay (see text for details). The bottom left panel shows the FRET efficiencies of the syn and anti conformations for the Rop dimers forming an A488-A647 pair (green and red dyes) or forming an A488-A594 pair (green and purple dyes). The formation of A488-A594 pairs would only occur if the original A488-A647 dimers dissociate during the structural rearrangement. When we performed this test, we observed the two FRET peaks corresponding to the syn and anti conformations of A488-A647 dimers, and no significant higher-FRET species that would indicate exchange with A594 monomers (as found in a separate control experiment, bottom right).
