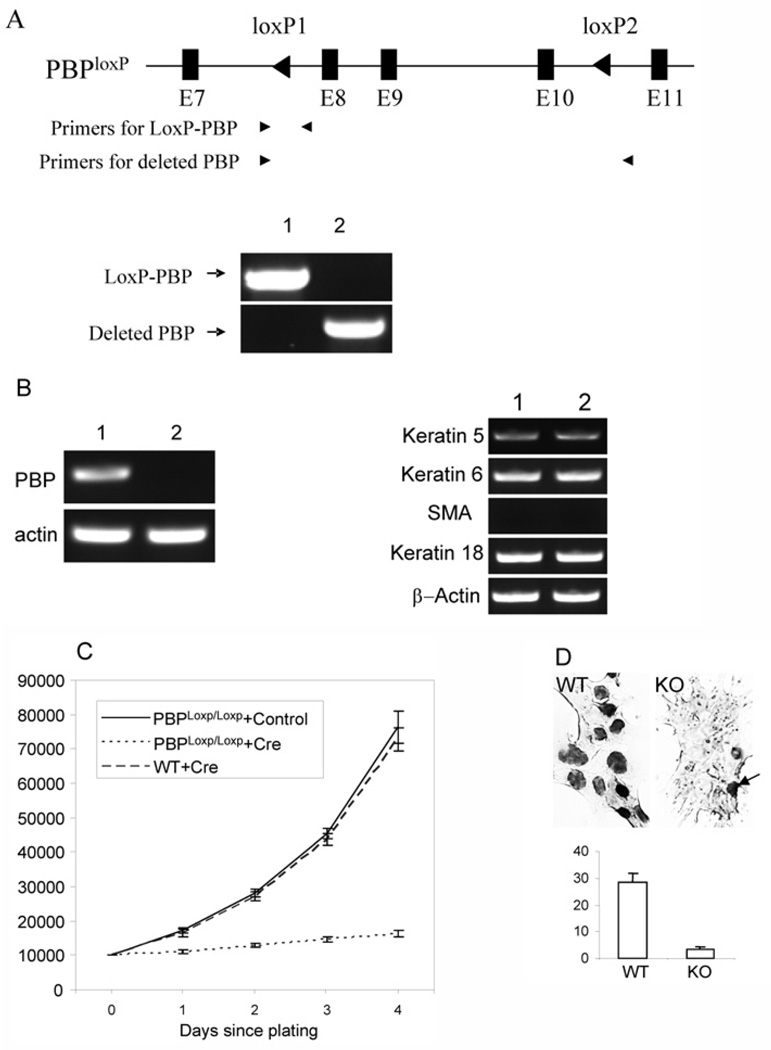
Figure 2. Requirement of PBP for the growth of Notch4-immortalized cells.
(A) Generation of PBP mutation. Notch4-immortalized cells with PBPLoxP/LoxP genotype were infected with retroviral Cre-ERtam or control viruses. After selection with puromycin and treatment with tamoxifen for 2 days, genomic DNA was prepared. PCR was performed with primers specific for the LoxP-PBP transgene (lane 1, +control; lane 2, +Cre-ERtam) and the deleted PBP gene. (B) Left panel: Loss of PBP mRNA expression in cells infected with retroviral Cre-ERtam. Lane 1, +control; lane 2, +Cre-ERtam. RT–PCR was performed to examine PBP expression. Right panel: the expression of markers for cell lineages was not altered due to loss of PBP expression, as revealed by RT–PCR. Lane 1, wild-type cells; lane 2, PBP-deficient cells. (C) Growth inhibition of PBP-deficient cells. PBPLoxP/LoxP Notch4-immortalized cells infected with retroviral Cre-ERtam or control viruses or wild-type Notch4-immortalized cells infected with retroviral Cre-ERtam were treated with tamoxifen for 2 days and then plated at 1×104 cells per well. Viable cells (y axis) were counted by Trypan Blue staining at the indicated time points after initial seeding. The proliferation of PBP-deficient cells was markedly inhibited (P < 0.001). (D) Markedly decreased nuclei involved in DNA synthesis in PBP-deficient cells, as revealed by BrdU labelling (top panel). WT, wild-type cells (PBPLoxP/LoxP cells infected with control viruses); KO, PBP-deficient cells. The arrow points to one of the few BrdU-labelled cells in the PBP knockout cells. Bottom panel: Quantification of BrdU staining in wild-type (WT) and PBP-deficient (KO) cells.