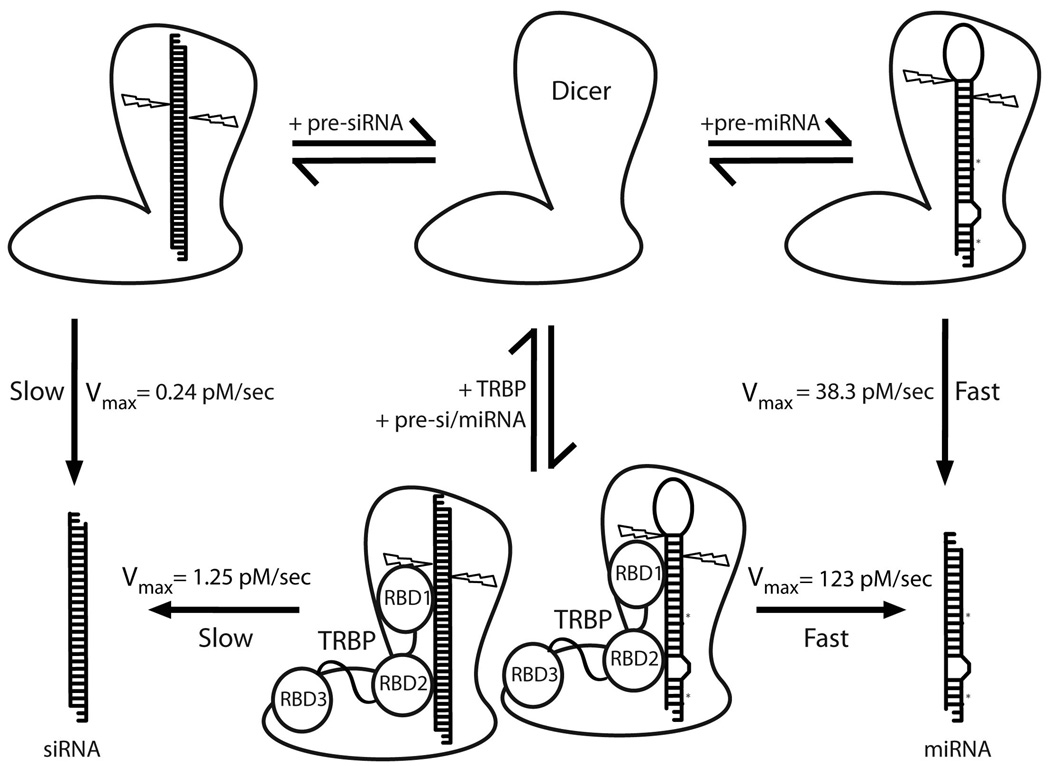
Figure 4.
Model depicting Dicer’s substrate selectivity and the effect of TRBP on dicing kinetics. Cartoon representatives of Dicer and Dicer-TRBP are based on a low resolution structure determined by electron microscopy 8. The ~100 fold difference in Vmax for pre-miRNA and pre-siRNA dicing could possibly reflect a difference in the loading efficacy, which in turn may be a function of the nature of the substrate. The N-terminal helicase domain of Dicer is known to bind the C-terminal dsRBD3 of TRBP, which in turn may mediate protein-nucleic acid interactions between the substrate RNA and one or both of the other dsRBDs of TRBP, thus stabilizing the enzyme-substrate complex. This could explain the 4–5 fold increase in activity of Dicer upon binding TRBP.