Figure 2.
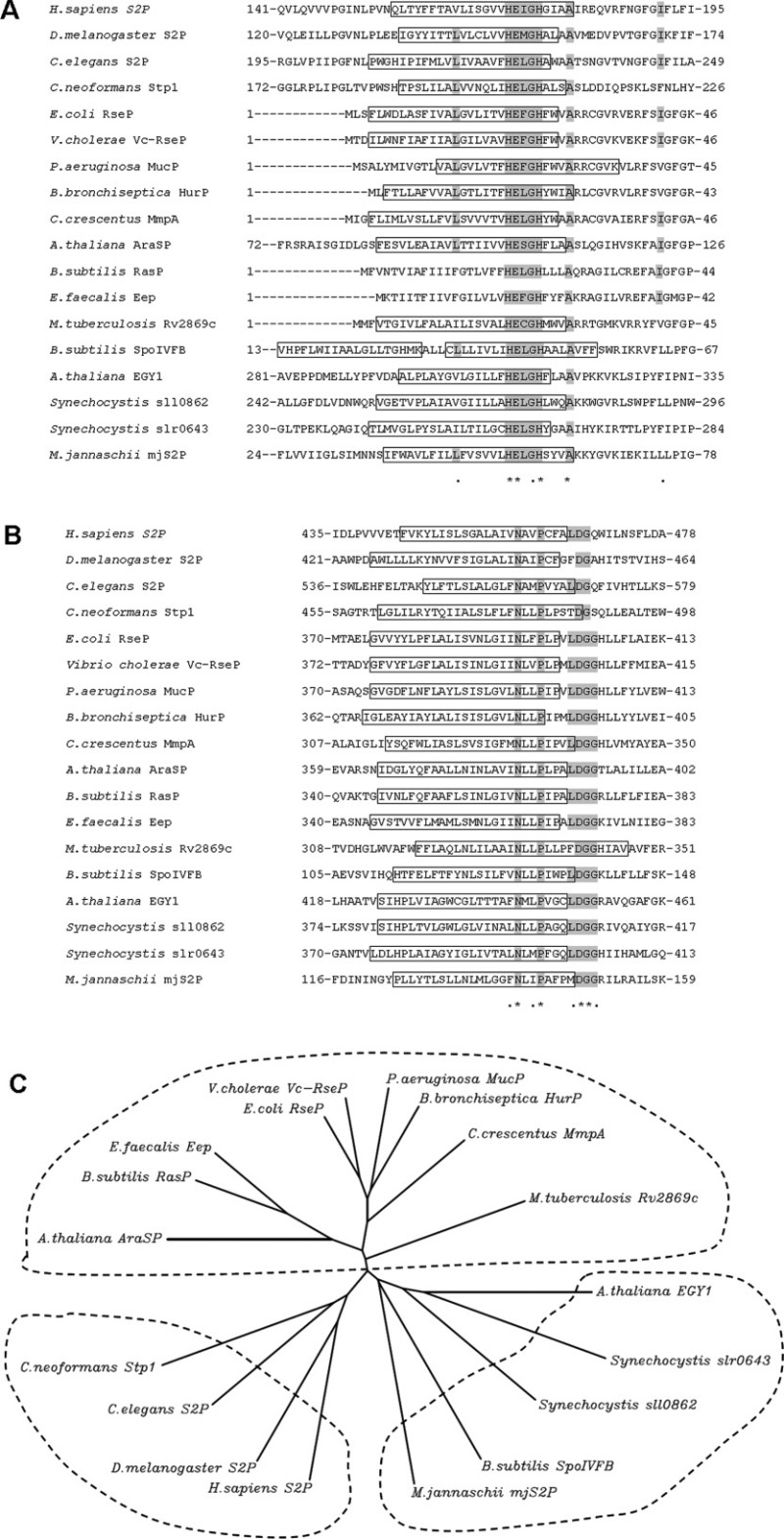
Sequence alignment and relationship among S2P homologs. The sequences of 18 characterized S2P homologs are presented. Only the core TM domains containing the HEXXH and NPDG motifs are shown. A: Sequence alignment in the vicinity of the HEXXH motif. The presumed transmembrane helix (TM) is boxed and the consensus amimo acids are highlighted in gray. B: Sequence alignment for the NPDG motif. C: Unrooted dendrogram of S2P homologs based on the alignment of the HEXXH and NPDG motifs. TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) and PSIPRED Protein Structure Prediction Server (http://bioinf.cs.ucl.ac.uk/psipred/) were used to predict TM helix. The unrooted dendrogram was generated by CLUSTALW (http://align.genome.jp/). The aligned sequences are: H.sapiens S2P (GI:6016601), D. melanogaster S2P (GI:19922044), C. elegans S2P (GI:22859116), C. neoformans Stp1 (BROAD ID: CNAG_05742), A. thaliana EGY1 (GI:15238440), Synechocystis sp. PCC6803 sll0862 (GI:16330216), Synechocystis sp. PCC6803 slr0643 (GI:16331565), A. thaliana AraSP (GI:18402981), E.coli RseP (GI:16128169), V. cholerae Vc-RseP (GI:20978850), P. aeruginosa MucP (GI:146448760), B. bronchiseptica HurP (GI:33601589), C. crescentus MmpA (GI:20978837), B. subtilis RasP (GI:20978800), E. faecalis Eep (GI: 256853722), M. tuberculosis Rv2869c (GI:20978863), B. subtilis SpoIVFB (GI:16079849), M. jannaschii mjS2P (GI:2499926).
