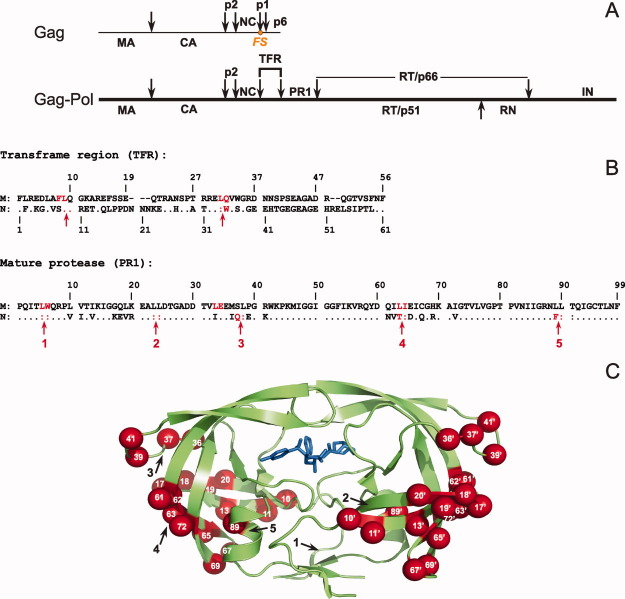
Figure 1.
A: Structural organization of Gag-Pol polyprotein in HIV-1. Arrows indicate the specific sites of cleavage by the viral protease. The protease is flanked at its N- and C-termini by the transframe region (TFR) and the reverse transcriptase, respectively. Nomenclature of HIV-1 proteins is according to Leis et al. 9; MA, matrix; CA, capsid; PR, protease; NC, nucleocapsid; RT, reverse transcriptase; RN, Rnase H; IN, integrase. For clarity, we denote mature proteases corresponding to Groups M and N as PR1M and PR1N. FS denotes the location of the frameshift site required for producing Gag-Pol. 6 B: Sequence alignment of TFR-PR1 domains derived from the Gag-Pol polyprotein of Groups M and N of HIV-1. The numbering system is relative to the N-terminus of TFR and the mature protease. The complete sequence for Group M is shown for reference, with dashes representing residues absent in TFR of Group M but present in the longer TFR of Group N. Dots (and colons at cleavage sites) indicate amino acids that are identical in the Group M and N sequences. Residues shown in red are major cleavage sites for TFR-PR1 autoprocessing (within TFR), and for the autoproteolysis of the mature PR (within PR), observed upon its release from the precursor. C: Sites of polymorphic substitutions mapped on the PR1N dimer described in this study. PR1N is shown as a green cartoon representation bound to inhibitor DRV, shown in blue sticks. The polymorphic substitutions are colored as red spheres on both the monomers and residue positions are labeled in white. Black arrows indicate the locations of the autoproteolytic sites shown in B. An interactive view is available in the electronic version of this article. PRO486 Figure 1