Figure 3.
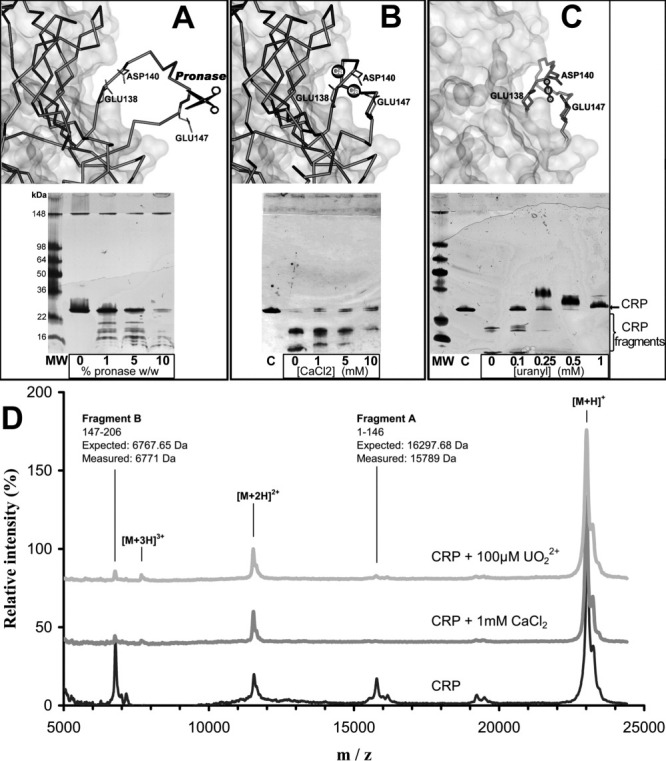
Conformational sensitivity of CRP to digestion with pronase. A: Top: without bound calcium atoms, the CRP138–148 loop is largely exposed to solvent (IGNH chain D). Bottom: electrophoresis gel of CRP (200 μg/mL) digestion at a fixed CaCl2 concentration (1 mM) and an increasing concentration of pronase, which promotes CRP (23 kDa) digestion. B: Top: in the presence of two bound calcium atoms, the CRP138–148 loop is less exposed to the solvent (1GNH chain C). Bottom: electrophoresis gel of CRP (200 μg/mL) digestion at a fixed concentration of pronase (5%W/W) and an increasing concentration of CaCl2. Calcium precludes CRP digestion by pronase41. C: Top: predicted binding site geometry of chelated  geometry on CRP chain C (IGNH). The side chains responsible for calcium chelation in CRP are also involved in
geometry on CRP chain C (IGNH). The side chains responsible for calcium chelation in CRP are also involved in  binding. Bottom: protection against pronase digestion of CRP (200 μg/mL) with increasing concentrations of the uranyl ion. Electrophoresis results represent at least two independent silver staining experiments. D: MALDI-TOF mass spectra of CRP. CRP was incubated without metal (black), with 1 mM CaCl2 (dark grey) and 100 μM
binding. Bottom: protection against pronase digestion of CRP (200 μg/mL) with increasing concentrations of the uranyl ion. Electrophoresis results represent at least two independent silver staining experiments. D: MALDI-TOF mass spectra of CRP. CRP was incubated without metal (black), with 1 mM CaCl2 (dark grey) and 100 μM  (light grey) prior to pronase digestion. Relative intensity was set at 100% for the [M+H]+ peak. [M+H]+, [M+2H]2+, and [M+3H]3+ are assigned to mono, doubly, and triply charged intact CRP peaks, fragments A and B are pronase digestion products. Calcium and
(light grey) prior to pronase digestion. Relative intensity was set at 100% for the [M+H]+ peak. [M+H]+, [M+2H]2+, and [M+3H]3+ are assigned to mono, doubly, and triply charged intact CRP peaks, fragments A and B are pronase digestion products. Calcium and  spectra show very similar peak distribution, with near disappearance of digestion products in agreement with electrophoresis results, implying that the conformation of the metal-bound CRP loop is similar for both metals.
spectra show very similar peak distribution, with near disappearance of digestion products in agreement with electrophoresis results, implying that the conformation of the metal-bound CRP loop is similar for both metals.
