Figure 1.
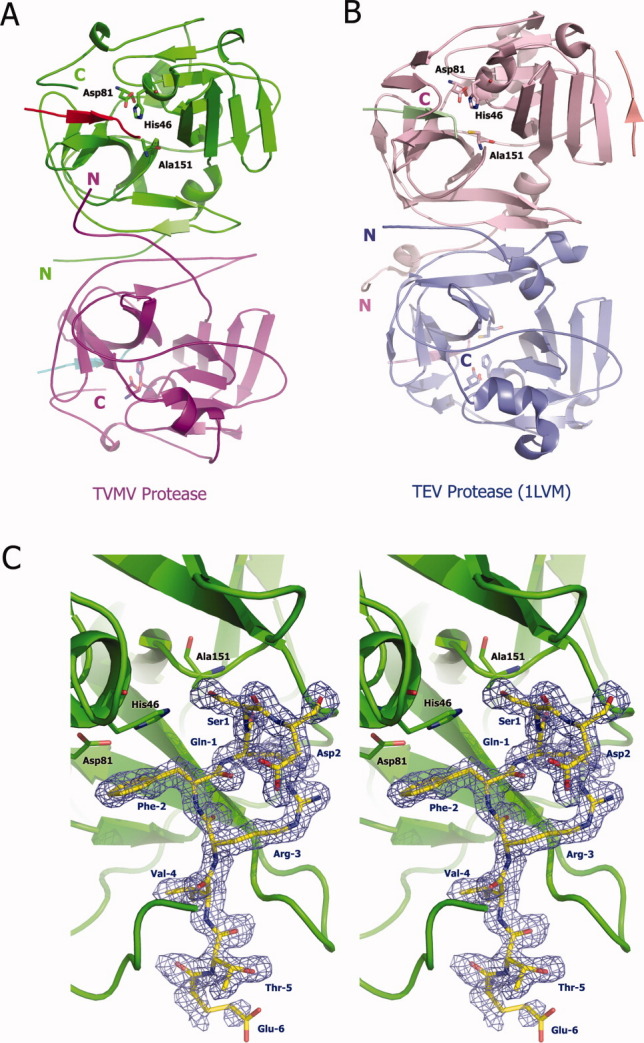
Three-dimensional structure of TVMV protease. (A) Overall structure of TVMV protease in one asymmetric unit. Chain A (1–217, purple) and Chain B (3–216, green) bound to peptide substrates Chain C (residues −6 to 1, cyan) and Chain D (residues −6 to 2, red), respectively. The N- and C-termini are labeled with the letters N and C. The catalytic triad residues are shown in ball-and-stick representation. (B) Crystal structure of TEV protease (PDB ID: 1LVM) viewed from the same perspective. Chains A to E are colored in pink, blue, green, pale pink, and salmon, respectively. (C) Stereoview of the peptide substrate (Chain D, yellow) bound to inactive TVMV protease (Chain B, green). The catalytic triad residues [H46, D81, and A151 (normally C151 in wild-type TVMV protease)] are shown in ball-and-stick representation. The peptide substrate is also displayed in a ball-and-stick format and covered by an omit map contoured at 1.0σ. The composite omit map was calculated at 1.7 Å resolution by CNS,13 with an omit rations of 7.5%.
