Abstract
Transcription factor, Jun dimerization protein 2 (JDP2), binds directly to histones and DNAs and then inhibits the p300-mediated acetylation both of core histones and of reconstituted nucleosomes that contain JDP2 recognition DNA sequences. JDP2 plays a key role as a repressor of adipocyte differentiation by regulation of the expression of the gene C/EBPδ via inhibition of histone acetylation. Moreover, JDP2-deficient mouse embryonic fibroblasts (JDP2−/− MEFs) are resistant to replicative senescence. JDP2 inhibits the recruitment of polycomb repressive complexes (PRC1 and PRC2) to the promoter of the gene encoding p16Ink4a, resulting from the inhibition of methylation of lysine 27 of histone H3 (H3K27). Therefore, it seems that chromatin-remodeling factors, including the PRC complex controlled by JDP2, may be important players in the senescence program. The novel mechanisms that underline the action of JDP2 in inducing cellular senescence and suppressing adipocyte differentiation are reviewed.
1. Introduction
The structure of chromatin, which influences numerous DNA-associated phenomena, such as transcription, replication, recombination, and repair, is controlled by a complex combination of histone modifications, ATP-dependent chromatin-remodeling enzymes, and nucleosome-assembly factors [1, 2]. The modification of histones such as acetylation, phosphorylation, methylation, ubiquitination, sumoylation, and ADP-ribosylation can regulate the gene expression [1–4]. The chromatin consists of structural units known as nucleosomes. Each nucleosome consists of two histone H2A-H2B dimers, a histone (H3-H4)2 tetramer, and DNA that is wrapped around the resultant histone octamers. During chromatin assembly, a histone (H3-H4)2 tetramer is formed before the two heterodimers of histones H2A and H2B are incorporated to form a nucleosome [5, 6]. The regulation of transcription is associated with alterations in chromatin structure that include histone modifications and changes in nucleosome structure [7–10]. Compaction of the chromatin and organization of nuclesomes represent a barrier that has to be overcome prior to the activation of transcription. The N-terminal histone tails that protrude from nucleosomes do not play a significant role in nuclesome formation but, rather, they appear to act as docking sites for other proteins and protein complexes to regulate chromatin compaction [9].
The structure of chromatin changes to allow greater accessibility by transcription factors when a gene is to be activated [10]. It has been suggested that the change to a more accessible state not only involves the modification of histones and alterations in nucleosomal arrays but also results from changes in nucleosome integrity that are due to displacement of histones [11]. Furthermore, it has been demonstrated that histone chaperones play a critical role in these processes [12–15]. Thus, it is tempting to speculate that histone chaperones might be important for the compaction of chromatin, and it is now important to determine whether certain corepressors of transcription might influence the deposition and assembly of nucleosomes through the regulation of histone-chaperone activity. The transcription factor Jun dimerization protein 2 (JDP2) is a member of AP-1 family that binds to both AP-1 site and cAMP responsive element (CRE) site in various cis-elements of the target genes. It is generally accepted that the transcription factors have DNA-binding activities and then control their transcriptional activities by DNA binding. However, we found that JDP2 has not only DNA-binding activity but also histone-binding activity. Moreover, JDP2 bound the nucleosome in both DNA sequence-dependent or -independent manner. JDP2 also has the nucleosome assembly activities and the activities of inhibition of histone acetyltransferase and histone methyltransferase. Here, we describe the role of JDP2 on the adipocyte differentiation and replicative senescence in mouse embryonic fibroblasts through histone modification.
2. JDP2 Regulates the AP-1-Mediated Activation of Transcription
JDP2 has been identified as a binding partner of c-Jun in yeast two-hybrid screening experiments, based on the recruitment of the SOS system [16]. JDP2 forms heterodimers with c-Jun and represses the AP-1-mediated activation of transcription [16]. Similarly, JDP2 was isolated by yeast two-hybrid screening with activation transcription factor-2 (ATF-2) as the “bait” [17]. JDP2 was also shown to associate with both the CCAT/enhancer-binding protein gamma (C/EBPγ) [18] and the progesterone receptor [19]. JDP2 is expressed in many cell lines and represses the transcriptional activity of AP-1 [20]. Moreover, JDP2 is rapidly phosphorylated at threonine residue 148 when cells are exposed to UV irradiation, oxidative stress, or inhibitor-induced depressed levels of translation by c-Jun NH(2)-terminal kinase (JNK) [21]. Although a novel JNK-docking domain is necessary for the activated kinase (MAPK) p38-mediated phosphorylation of JDP2 at threonine residue 148, this domain is not sufficient for this process [22]. JDP2 binds to both cAMP-responsive element (CRE) and TPA-responsive elements (TREs) on DNA as a homodimer and as a heterodimer with ATF-2 and members of the Jun family, respectively [16, 17]. JDP2 encoded an 18 kDa protein that is able to homodimerize as well as to form heterodimers with other AP-1 members, such as c-Jun, JunB, JunD, and ATF-2, and members of the C/EBP family, C/EBPγ and C/EBP homologous protein 10 (CHOP 10) [16, 17] as well as Interferon regulatory factor- (IRF-) binding protein 1 [23]. Dimerization occurs through a conserved leucine zipper domain found all members of the AP-1 family. A basic domain located adjacent to the leucine zipper dimerization motif is responsible for the direct association with TRE and CRE [16, 17]. In vitro study using the purified JDP2 protein showed that JDP2 forms the homotrimer but not the homodimer in our biochemical condition (unpublished data). JDP2 inhibits UV-induced apoptosis by suppressing the transcription of the p53 gene [24]. Given the roles of AP-1 in cellular transformation and the reported repression of Jun- and ATF-2-mediated transcription by JDP2, we have demonstrated that JDP2 inhibits the oncogenic transformation of chicken embryonic fibroblasts [25]. JDP2 also modulates the expression of cyclin D1 and p21, which have opposing effects on cell-cycle progression. JDP2 interferes with the progression of the cell cycle by reducing the levels of cyclin D1 and at the same time increases the expression of p21 [26, 27]. The forced expression of JDP2 promotes the myogenic differentiation of C2C12 cells, which is accompanied by the formation of C2 myotubes and the strong expression of major myogenic markers. Moreover, the ectopic expression of JDP2 in rhabdomyosarcoma cells induces incomplete myogenesis and the incomplete formation of myotubes [27]. These cells become committed to differentiation via the p38-MAPK pathway [21, 22]. A similar enhancement of cell differentiation was reported during the induction of osteoclast formation by the receptor activator of the nuclear factor kB (NFkB) ligand (RANKL) [28]. Unlike other members of the AP-1 family, the levels of JDP2 remain constant in response to a large variety of stimuli, such as UV, irradiation, and retinoic acid (RA), which affect the levels of other factors involved in cell-cycle control. The induction of JDP2 expression was only observed during the differentiation of F9 cells to muscle cells and osteoclasts. Therefore, JDP2 may provide a threshold for exit from the cell cycle and a commitment to differentiation (Figure 1). Further studies of the regulation of the cell cycle and the differentiation of cells induced by JDP2 should be very instructive. It is also interesting that JDP2 is one of the candidate oncoproteins that collaborate in the oncogenesis associated with the loss of p27 as the result of insertional mutations [29]. Recent study of tumor cells demonstrated that JDP2 was a tumor suppressor [30]. We have also found that JDP2 is a repressor of the activation of transcription via AP-1 and a negative regulator of the retinoic acid- (RA-) induced differentiation of mouse embryonic F9 cells and adipocyte differentiation [31–33].
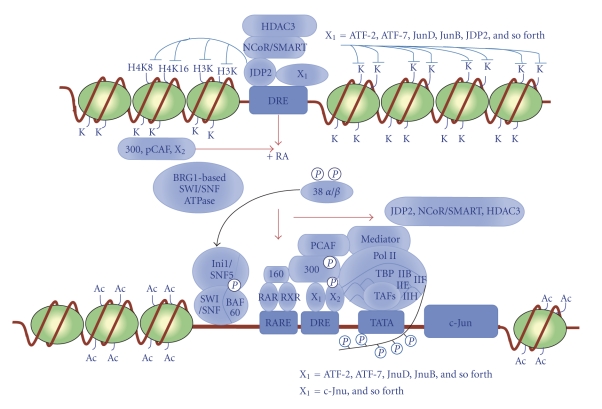
Figure 1.
Schematic representation of the signal pathways of retinoic acid- (RA-) induced (RA-induced) differentiation of mouse embryonic carcinoma F9 cells. At the undifferentiation stage of F9 cells, HDAC3, NcoR/SMART, and JDP2 were recruited on the DRE (differentiation response element) in the promoter region of the c-jun gene to induce the heterochromatin. In the response to retinoic acid (RA), the signals of mitogen-activated kinase (MAPK; phosphorylated (p38(α/β)), BRG1-based SWI/SNF ATPase complex, Ini1/Snf5/SWI/SNF/BAF60 complex, p160 hormone coactivator (RARE/RXRE binding), and p300/PCAF complex were recruited to the DRE element of the c-Jun promoter and then Mediator, Pol II complex (IIB, IIF, IIE, IIH complex), TBP complex, and some TAF complex (ATF12, TAF 4 etc), and recruited and Pol II complex is elongated with phosphorylation of the carboxy terminal domain (CTD) of Pol II, and then c-Jun genes are finally activated. Ac, acetylated residues; K, lysine residues of histone; H3K, lysine residues of histone H3; H4K8, lysine residue at position 8 of histone H4; H4K16, lysine residue at position 16 of histone H4; X1 = ATF-2, ATF-7. JunD, JunB, JDP2, and so forth; X2 = c-Jun and so forth.
3. JDP2 Inhibits Histone Acetyltransferase (HAT) Activity
We have reported previously that JDP2 represses the transactivation mediated by p300 [32]. Both p300 and ATF-2 have HAT activity [34, 35]. It was recently shown that p300 acetylates ATF-2 protein in vitro at lysine residues 357 and 374 and that ATF-2 is essential for the acetylation of histones H4 and H2B in vivo [36, 37]. We found that acetylation by p300 is inhibited in a dose-dependent manner by JDP2, when added exogenously. We also found that JDP2 was not acetylated by p300 under our experimental conditions. The inhibitory effect of JDP2 was detected on histone acetylation induced by p300, CREB-binding protein (CBP), p300/CBP-associated protein (PCAF), and general control nonrepressive 5 (GCN5). The overexpression of JDP2 apparently represses the RA-induced acetylation of lysines 8 and 16 of histone H4 and some amino terminal lysine residues of histone H3.
4. JDP2 Has Intrinsic Nucleosome-Assembly Activity In Vitro
The template activation factor-1β protein (TAF-Iβ protein), which is a component of the inhibition of histone acetylase complex (INHAT complex) identified by Seo et al. [38, 39], is a histone chaperone that binds directly to core histones and facilitates the assembly of nucleosomes in vitro. JDP2 interacted directly with all the core histones tested and inhibited the p300-mediated acetylation of those histones. To our surprise, JDP2 also introduced supercoils into circular DNA in the presence of core histones, to levels similar to those observed for yeast CCG1-interacting factor 1 protein (yCia1p) and CCG1-interactiing factor (CIA1). Therefore, JDP2 appears to have significant histone chaperone activity in vitro [32]. We have also shown that the HAT-inhibitory activity of JDP2 is involved, to some extent, in the repression of transcription by JDP2, whereas the maximal capacity of JDP2 to suppress the RA-mediated activation of the c-Jun promoter [32, Figure 1] and to suppress the adipocyte differentiation [33] requires the recruitment of histone deacetylases (HDACs).
5. JDP2 Suppresses Adipocyte Differentiation
JDP2 has been shown to play a role in the cellular differentiation of skeletal muscle, osteoclasts, adipocytes [27, 33], and F9 cells [31]. It has been reported that JDP2 has an activity of tumor suppressor using prostate cancer cells [30]. Recently, we also found that JDP2-deficient mouse fibroblasts are resistant to replicative senescence [40]. Finally general inhibition of the AP-1 complex by expression of JDP2 specifically in the heart correlates with the induction of atrial dilatation [41]. In mammals, the strict control of adipocyte development, the mass of adipose tissue, the insulin sensitivity of adipocycles, and the appropriate metabolism of glucose and lipids are critical to the maintenance of energy homeostasis [42, 43]. Adipogenesis, namely, the process whereby hormonal stimuli induce the differentiation of fibroblasts or mesenchymal cells to adipocytes, requires the organized and controlled expression of a cascade of transcription factors and the modification of the chromatin within preadipocytes [44–47]. The factors involved in adipocyte differentiation include a nuclear receptor known as peroxisome proliferation-activated receptor gamma (PPARγ) and a group of C/EBPs [48, 49]. The rapid and transient induction of the expression of C/EBPβ and C/EBPδ is one of the earliest events in adipogenesis [46]. These transcription factors bind to specific sequences in the promoters of the C/EBPα gene and the PPARγ gene, inducing their expression, which, in turn, activates the full adipogenic program of gene expression [50–53]. Expression of PPARγ is also induced via a sterol-regulating element-binding protein-1c- (SREBP-1c-) dependent pathway [54]. Once both PPARγ2 and C/EBPα are activated, “cross-talk” between PPARγ and C/EBPα maintains the expression of each protein during adipocyte differentiation, even in the absence of C/EBPβ and C/EBPδ [55]. We generated JDP2 “knock-out” (KO) mice in order to study the activities of JDP2 in vivo, and we found that JDP2 plays a role as a repressor of adipocyte differentiation. We also found that JDP2 targeted an adipogenesis-related gene, C/EBPδ, and inhibited its expression via regulation of histone acetylation.
The C/EBPδ and C/EBPα mRNAs were overexpressed in Jdp2−/− MEFs during the initial stages of adipocyte differentiation, whereas C/EBPβ and PPARγ mRNAs were less affected by the absence of JDP2 [56]. C/EBPα is known to be the downstream target of C/EBPδ and C/EBPβ [57]; therefore, it can be speculated that the augmentation of expression of the C/EBP gene might have been due to indirect effects, which were probably caused by the enhanced expression of the C/EBPδ gene [33]. In fact, the recruitment of JDP2 to the C/EBPδ and C/EBPβ genes was detected, but there was no obvious recruitment to the C/EBPα gene. Thus, the target of JDP2 is possibly the C/EBPδ and C/EBPβ genes.
JDP2 was able to bind to the C/EBPδ promoter to repress the transcription of the C/EBPδ gene. Our JDP2-deficient mice did not have abundant adipose cells, in other words, they did not have a thick layer of fat tissue, perhaps because the mass of adipose tissue might be determined by more complex factors, such as cytokines and hormones, via as yet unknown mechanisms, and by energy status. We found that the adipose tissue of scapulae from young Jdp2−/− mice consisted mostly of white adipocytes, whereas the majority of cells were brown adipocytes in WT mice. This observation suggested that JDP2 might play a role in adipogenesis in vivo.
Recently, it has been reported that the activation of Wnt signalling inhibited the development of brown adipocytes by mediating the peroxisome proliferation-activated receptor gamma-1 coactivator-1-1α- (PGC1-1α)-) uncoupling protein1 (UCP1) cascade [48]. In addition to C/EBPs as shown in the present studies, JDP2 might be a target of such signal networks. Other report suggested that the absence of Rb switched the differentiation of mouse embryonic fibroblast (MEF) cells from white to brown adipocytes [58], suggesting that the differentiation of WT MEFs to adipocytes is a good model for white-adipocyte differentiation but not for brown-adipocyte differentiation. We also observed that PGC-1α marker of brown adipocyte was not induced after induction of adipocyte differentiation in Jdp2−/− MEFs and WT MEFs (unpublished). Then, we analyzed the levels of mRNA of PGC-1α in Adeno-JDP2-infected 3T3-L1 cells. 3T3-L1 cells are also known to be a model for white-adipocyte differentiation, however, we could detect small amount of PGC-1α RNA by real-time RT-PCR as the others reported elsewhere [59] probably because quite minor population of 3T3-L1 cells can differentiate to brown adipocyte. The expression of PGC-1α was increased after adipocyte differentiation, and the levels of expression were not remarkably different between Adeno-JDP2- and Adeno-lacZ -infected 3T3-L1 cells. So, we speculate that JDP2 might inhibit the differentiation of white adipocytes exclusively, but not brown adipocytes. In consequence, brown adipocytes are preferentially developed in WT mice scapulae, whereas white adipocytes appear in Jdp2−/− mice. However, further studies are required to clarify the role of JDP2 in the molecular mechanism that directs fibroblast and mesenchymal cells to differentiate into white or brown adipocytes.
6. JDP2 Controls Replicative Senescence
We analyzed the aging-dependent proliferation of MEFs from Jdp2−/− ice in the presence of environmental (20%) or low (3%) oxygen [40]. The Jdp2−/− MEFs continued to divide, even after six weeks, whereas the wild-type MEFs almost stopped proliferating and entered senescence under environmental oxygen. Conversely, neither the wild-type MEFs nor the Jdp2−/− MEFs succumbed to replicative senescence at lower oxidative stress. These results demonstrate that MEFs lacking JDP2 can escape from the irreversible growth arrest caused by environmental oxygen. The expressions of p16Ink4a and alternative reading frame (Arf) were repressed in aged Jdp2−/− MEFs (40 days) compared with their levels in wild-type MEFs. In 3% oxygen, at the equivalent time (40 days), wild-type MEFs expressed lower levels of p16Ink4a and Arf compared with those in 20% oxygen, whereas Jdp2−/− MEFs maintained low-level expression of p16Ink4a and Arf (Figure 3). These observations indicate that the aging-associated expression of p16Ink4a and Arf is dependent on oxygen stress and that JDP2 controls the expression of both p16Ink4a and Arf. We found no dramatic downregulation of the upstream repressors of p16Ink4a/Arf, B lymphocyte molony murine leukemia virus insertion region 1 homology (Bmi1), and enhancer of zesta homolog 2 (Ezh2), in the absence of JDP2, suggesting that JDP2 does not regulate their expression. Interestingly, JDP2 expression in wild-type MEFs increased in the presence of 20% oxygen, but not in the presence of 3% oxygen, suggesting that its expression depends on oxygenic stress and that accumulated JDP2 may play a role in the transcriptional activation of p16Ink4a/Arf (Figure 2). Studies based on ChIP have demonstrated that the methylation of H3K27 at the p16Ink4a/Arf locus was higher in Jdp2−/− MEFs than in wild-type MEFs and that the binding of polycomb repressor complex 1(PRC1) and polycomb repressor complex 2 (PRC2) to the p16Ink4a and Arf promoters was more efficient in Jdp2−/− MEFs than in wild-type MEFs. These observations suggest that, in the absence of JDP2, H3K27 is methylated by PRC2, and the p16Ink4a/Arf locus is silenced by PRC1, whereas the increased expression of JDP2 helps to release PRC1 and PRC2 from the p16Ink4a/Arf locus, thereby reducing H3K27 methylation. Our data demonstrate that JDP2 is an important factor regulating cellular senescence. The loss of JDP2 allows MEFs to escape senescence, and conversely, the overexpression of JDP2 induces cell-cycle arrest. The absence of JDP2 reduces the expression of both p16Ink4a and Arf, which inhibit cell-cycle progression.
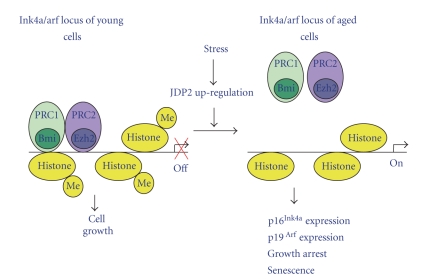
Figure 3.
Proposed model of the epigenetic regulation of the expression of the genes for p16Ink4a and Arf by JDP2. The exposure of young MEF primary cells to aging stress leads to the accumulation of JDP2. JDP2 binds to histones and inhibits the methylation of H3K27 at the p16Ink4a/Arf locus. As a result, PRC-1 and PRC-2 fail to form stable repressive complexes and are released from the locus. The consequent expression of p16Ink4a and Arf in the aged cells leads to growth arrest and senescence stage.
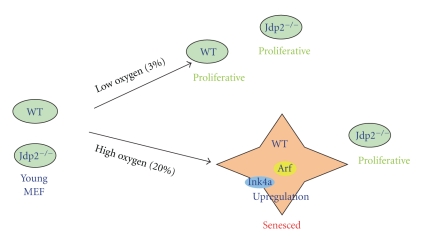
Figure 2.
Oxydative stress controls the cell proliferation of mouse MEF cells through the epigenetic regulation of the p16Ink4a/Arf locus by JDP2. Young MEF primary cells exposed to oxidative stress (20%) accumulate JDP2. In the presence of JDP2, PRC1, and PRC2 dissociate from the p16Ink4a/Arf locus, and histone H3 on the promoter is demethylated. Finally, p16Ink4a and Arf are upregulated to express and then aged cells senescence. In the absence of JDP2, MEF cells do not enter the senescence stage and proliferate well. In the lower oxygen (3%), both WT and Jdp2−/− MEF cells are not senesced but proliferated.
6.1. p16Ink4a and the Rb Pathway
Genes that are essential for cell-cycle progression are transcribed at the beginning of G1 phase by transcription factors of the E2F family. E2F is controlled by Rb family of proteins, pRb, p107, and p130 [60, 61]. Early in G1, unphosphorylated Rb proteins bind to the E2F family of proteins and inactivate their function [62, 63]. During G1, the Rb proteins are inactivated by phosphorylation by Cdk4/6-cyclinD complexes, thereby allowing the transcription of E2F-dependent gene, including cyclin E. Upregulated cyclin E forms a complex with cdk2, that mediates the hyperphosphorylation of the Rb proteins, an essential requirement for the G1/S transition. p16Ink4a is an allosteric inhibitor of cdk4/6. Binding to p16Ink4achanges the conformation of cdk4/6, which prevents its interaction with cyclin D [64, 65]. Therefore, p16Ink4a acts as an inhibitor of the cell cycle at G1 by modulating the Rb pathway. p16Ink4a is often lost in a variety of human malignancies, including glioblastoma, melanoma, and pancreatic adenocarcinoma [66]. In contrast, the upregulation of p16Ink4a induces the cell-cycle arrest and senescence [65, 66].
6.2. Arf and the p53 Pathway
p53 is known to mediate cell-cycle arrest, in G1 and G2, and apoptosis. A number of downstream targets of p53 are involved in these processes, including p21Cip/waf1 in G1 arrest [67], 14-3-3 sigma and growth arrest and DNA damage inducible gene 45 (GADD45) in G2 arrest [68, 69], and p21, Bax, PI-3 upregulated modulator of apoptosis (PUMA), Fas/Apo1, and Killer/DR5 in apoptosis [70–74]. p53 is regulated at the levels of protein stability and activity, and to some extent at transcription and translation [75, 76]. In unstressed cells, p53 protein levels are very low because its degradation is mediated by the E3 ubiquitin ligase activity of murine double minute 2 (MDM2), which targets p53 for ubiquitin-dependent proteolysis [77]. MDM2 is a transcriptional target of p53, so p53 directly activates the expression of its own negative regulator, producing a potent negative feedback regulatory loop [78]. There are several stress-responsive kinases, which, by phosphorylating p53, inhibit its degradation by MDM2 and increase its transcriptional activity [79–81]. DNA damage rapidly activates the ataxia telangiectasia mutated (ATM) and ataxia elangiectasia-related (ATR) proteins, which phosphorylate the checkpoint kinases 1 and 2 (Chk1 and Chk2), which in turn propagate the signal to downstream effectors such as p53 [82, 83]. Both Chk1 and Chk2 phosphorylate p53 at Ser 20, which prevents the efficient recruitment of MDM2. Thus, p53 is stabilized and its expression level is increased in response to stress signaling. Arf is predominantly localized in the nucleoli and is stabilized by binding to nucleophosmin. In response to stress signaling, Arf is released from nucleophosmin and translocates to the nucleoplasm, where it interacts with MDM2, inhibits its E3 ubiquitin ligase activity, and blocks the nucleocytoplasmic shuttling of the MDM2-p53 complex. Therefore, the consequences of the activation of Arf are the stabilization and activation of p53 [84, 85].
6.3. Senescence and Aging in Human and Mouse
Cellular senescence appears to be related to organismal aging. Cellular senescence involves processes that include telomere shortening, the accumulation of DNA damage, and the activation of the p16Ink4a/Arf locus. The contributions of these factors to senescence seem to differ in humans and mice. Cultured mouse fibroblasts undergo senescence even when they have long telomeres and high telomerase activity. Senescence is abrogated by the loss of the p16Ink4a/Arf locus [86]. In human cell cultures, the ectopic expression of telomerase is sufficient to overcome senescence by maintaining the length of the telomeres [87]. In mice, the maintenance of telomere length is important because telomerase deficiency shortens their lifespan and leads to premature aging [88–92]. The age-dependent accumulation of INK4A has been observed in the human kidney and skin [91], as well as in the majority of mouse tissues [92]. In oncogene-induced senescence, there is in vivo evidence that Arf is the important factor in the activation of p53 tumor suppression [91]. However, another study has shown that components of the DNA-damage-signaling cascade, including ATM and Chk2, are critical for the activation of p53 in response to oncogenic signals [93]. These differences between humans and mice could be attributable to species specificity and/or experimental conditions. Cellular senescence appears to be related to organismal aging because the same processes appear to be involved. Genetic variants of the p16Ink4a/Arf locus are linked to age-associated disorders, such as general frailty, heart failure, and type 2 diabetes [94–99]. Mutations in telomerase or in proteins that affect telomerase activity are linked to premature human aging syndromes, including congenital dyskeratosis and aplastic anemia [100]. There are increases in DNA mutations, DNA oxidation, and chromosome loss during organismal aging. It seems reasonable to assume that all three factors, the activation of the p16Ink4a/Arf locus, telomere shortening, and the accumulation of DNA damage, have cooperative effects on aging in physiological situations. Understanding the mechanisms of cellular senescence is currently of wide interest, and it is important that we identify new components of this process, such as JDP2.
7. Interaction of Transcription Factors with Histones and Nucleosomes
Similar to JDP2, other transcription factors or chromatin regulators are reported to have the binding affinities to histones and/or nucleosome. Transcription factor IIIA (TFIIIA) is a 40-kDa protein, with nine zinc finger domains, that binds specifically to the internal promoter of the gene for 5S RNA and to the N-terminal tail domains of histones H3 and H4, but not those of H2A and/or H2B, and directly modulates the ability of TFIIIA to bind nucleosomal DNA [101, 102].
A new class of HAT-regulatory proteins has been identified. These proteins block HAT activity via binding to and masking of the histone themselves. This class includes the subunits of INHAT complex; TAF-1α, TAF-1β, and pp32 as well as ataxin 3; silencing mediator or retinoid receptor corepressor (SMART)/nuclear hormone receptor corepressor (NcoR); proline-glutamic acid- and leucine-rich protein 1 (PELP1) [103–107]. Thanatos-associated protein 7 (THAP7) is known to associate with TAF-1β and to repress transcription by inhibition histone acetylation [108, 109]. A novel INHAT repressor (NIR) binds directly to nucleosomes and core histones and prevents acetylation by histone acetyltransferases, thus acting as a bona fide INHAT [110]. PU.1, a member of the Ets family of oncoproteins, inhibits CBP-mediated acetylation of globin transcription factor-1 (GATA-1) and erythroid Krüppel-like factor (EKLF), as well as of histones, and disrupts acetylation-dependent transcriptional events [111]. Moreover, PU.1 recruits the retinoblastoma tumor-suppressor protein, a histone methyltransferase (HMT) Suv39H, and heterochromatin protein 1α (Hp1α) [112]. Similarly, proliferating cell nuclear antigen binds to p300 to inhibit its HAT activity in vitro and to block HAT-dependent transcription in vivo [113]. TAF-1β and pp32 have also been shown to interact with estrogen receptor (ERα), to inhibit the ERα-mediated activation of transcription, and to inhibit the activation of ERα acetylation by p300 [106, 107]. Moreover, TAF-1β also interacts with the Sp-1 transcription factor and with KLF5, negatively regulating the binding to DNA and activation of transcription by these factors [114, 115]. Transcription factor NF-kB p50 can accommodate distorted, bent DNA within the nucleosome [116], while DNA-binding domain (DBD) of c-Myb binds to the N-terminal tails of H3 and H3.3, and binding of c-Myb facilitates acetylation of histone tails [117]. Moreover, a kinetochore-null protein (KNL-2) with a c-Myb-like DNA-binding domain is specifically required for loading of centromere-specific variants of histone H3 (centromere protein A; CENP-A) in both nematodes and mammalian cells [118]. The recruitment of repressive macroH2A nucleosomes requires direct interactions between ATF-2 bound to a nearby AP-1 site and macroH2A, and recruitment is regulated by DNA-induced protein allostery [119]. Thus, the abovementioned sequence-specific DNA-binding factors might regulate transcription either via histone cores or tails, as well as via the structure of nucleosomes in conjunction with other proteins that bind to the chromatin. Indeed JDP2 is the first case to have both nuclesome assembly activity and DNA-binding activity [32] and inhibits the histone acetylation by p300/CBP [32, 33] as well as histone methylation [40] as described below.
8. Mechanism of JDP2-Mediated Regulation of C/EBPδ and p16Ink4a/p19
Differentiation and senescence are associated with dynamic changes in gene expression, which are regulated by chromatin remodeling. Here, we have shown that the expression of JDP2 regulated the differentiation competent genes and the senescence competent genes such as C/EBPδ and p16Ink4a and Arf in response to the induced hormone and accumulating oxidative stresses. In the case of suppression of adipocyte differentiation by JDP2, how might JDP2 inhibit transcription of the C/EBPδ gene? It seems possible that, in response to signals that lead to differentiation, the C/EBPδpromoter might recruit transcription factors, including the coactivator p300, that mediate the acetylation of histones associated with the C/EBPδ gene to stimulate transcription of this gene. In the presence of JDP2, the acetylation of histone is inhibited and the expression of the C/EBPδ gene is suppressed during adipocyte differentiation, even in the presence of differentiation-inducing signals. Decreased expression of C/EBPδ results in less effective differentiation to adipocytes, a conclusion that is consistent with a previous report that deletion of the C/EBPδ gene results in the impairment of adipocyte differentiation [59]. By contrast, HDAC3 was not recruited by JDP2 to the promoter region of the C/EBPβ gene, although the recruitment of HDAC3 to the C/EBPβ gene was slightly enhanced during differentiation. In addition, the acetylation of histone H3 on the C/EBPβ gene was less affected by differentiation-inducing stimuli. These observations suggest that histone acetylation might not play an important role in the transcriptional regulation of the C/EBPβ, gene and the HDAC3 might not be involved in such regulation. This hypothesis explains why the presence of JDP2 on the promoter had no effect on transcription. In conclusion, we propose that JDP2 acts as a negative molecular-switch in some, but not all, types of differentiation via the regulation of the expression of specific genes in concert with histone deacetylase HDAC3, as shown in the present study of adipocyte differentiation.
In the case of induction of cellular senescence by JDP2, we propose a model; the accumulation of oxidative stress and/or other environmental stimuli during aging upregulate JDP2 expression in primary untransformed cells. Increased JDP2 helps to remove PRC1 and PRC2, which are responsible for the methylation of histone H3, from the p16Ink4a/Arf locus, leading to increased p16Ink4a and Arf expression and entry into the senescence (Figure 4). There is some evidence that JDP2 acts as a tumor suppressor; JDP2 inhibits the Ras-dependent transformation of NIH3T3 cells [29], and JDP2 gene disruptions are often found in the lymphomas induced by insertional mutagenesis caused by the Moloney murine leukemia virus in MYC/Runx2 transgenic mice [27]. Here, we suggest that JDP2 not only inhibits the transformation of cells but also plays a role in the induction of cell senescence. Both functions of JDP2 might be important for its role in inhibiting tumor formation. Our findings also provide new insights into the molecular mechanisms, by which senescence is induced in the context of the epigenetic regulation of the p16Ink4a/Arf locus. Recently, we reported that JDP2 regulates the expressions of cell-cycle regulators such as cyclin A2, cyclin E2, and p16Ink4a and also affects the expression of p53 and p21 protein [120]. Thus, it is evident that JDP2 control the expression of cell-cycle regulators to induce the cell-cycle arrest via p53-p21 pathway and RB-p16 pathway.
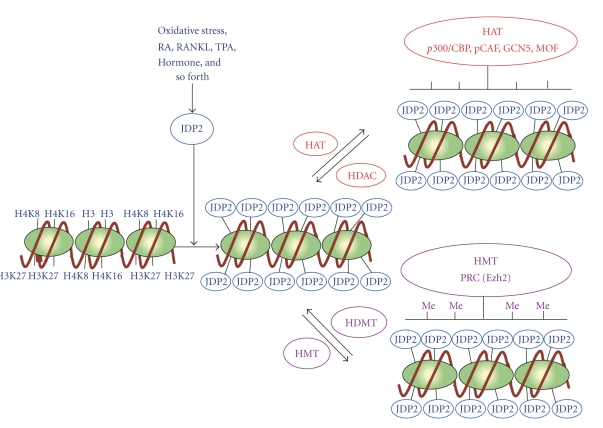
Figure 4.
Model for the epigenetic regulation by JDP2. During the exposure of cells with oxydative stress, retinoic acid (RA), RANKL, TPA, and adipocyte inducing hormones, the histone H3, H4K8, and H4K16 as well as H3K27 were masked by JDP2 proteins and prevent the attack of histone modification enzymes like HAT (p300/CBP, pCAF, GCN5, MOF, etc.) and HMT (Ezh2 etc) in the cellular senescence and cell differentiation. This is a novel mechanism of JDP2 to inhibit the histone modification. The gene locus is either the p16Ink4a/Arf locus or C/EBPδ locus.
9. Concluding Remarks
We characterized a sequence-specific DNA-binding protein as a nucleosome-assembly factor. Our finding should facilitate efforts to understand some aspects of nuclesome assembly and the remodeling of chromatin. Histone H3 seems to be recruited to target sites in a DNA synthesis-dependent manner via the interaction of chromatin assembly factor 1 (CAF-1) with proliferation cell nuclear antigen (PCNA); by contrast, a histone chaperone HIRA was reported to incorporate H3.3 in a DNA synthesis-independent manner [15]. The observation that histone H3.3 is found in several other subcomplex suggests that H3.3 might be recruited to different sites by different pathways. Identification of a gene-specific DNA-binding protein, namely JDP2, as a nucleosome-assembly factor suggests that H3.3 could be directly deposited at specific locations by site-specific DNA-binding proteins that also have histone-chaperone activity. It will be of interest to examine the binding preference of JDP2 for histone H3 as compared with H3.3.
Since JDP2 binds to core histone or nuclesome partially in DNA sequence-specific manner or histone subset-specific manner, the histone acetyltransferase or histone methytransferase may not access to the nucleosome in vitro (Figure 4). However, we are not sure the case of in vivo. Chromatin immunoprecipitation assay (ChIP assay) demonstrated that JDP2 inhibited at least the acetylation of histone H4K8 and H4K16 although we cannot determine other precise residues of histone H3 acetylation (31). Moreover, JDP2 associated with histone H3K27 and blocked the methylation of histones (40). Thus, we assume that the interaction of JDP2 with nucleosome is DNA sequence or histone modification specific, and thus, only certain restricted set of histone might be associated with JDP2 in vivo. Addressing these precise functions in the context of epigenesis helps us to understand how senescence and differentiation, in a broader context, are regulated.
10. Future Prospects
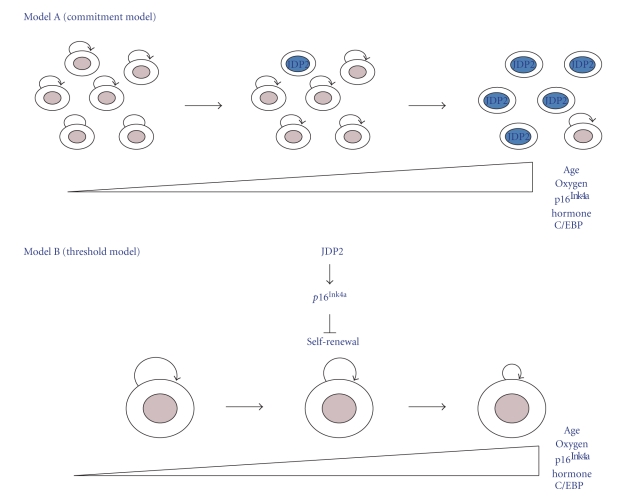
Understanding how JDP2 promotes aging or cell differentiation, whether by inducing cellular senescence or decreasing the frequency of cell-cycle entry, is an important issue. The data demonstrating the increase in expression of p16Ink4a or C/EBPδ with aging (or oxygen stress) or hormone induction can be reconciled with two different models like “commitment model” and “threshold model.” With aging or hormone induction, stochastic activation of p16Ink4a or C/EBPδ expression could occur on a cell-by-cell basis in self-renewing compartments to induce commitment (commitment model), or expression could increase simultaneously with the majority of cells of a self-renewing compartment (threshold model) (Figure 5). In the latter case, self-renewal would be impaired by p16Ink4a or C/EBPδ expression by decreased frequency of cell-cycle entry in the absence of senescence or cell differentiation.
Figure 5.
Model of role of JDP2 in cellular aging and cell differentiation. Self-renewing cells undergo repeated divisions, and p16Ink4aor C/EBPδ expression increases with age, oxygen, and hormone, as a consequence of undefined stimuli. (a) Commitment model: p16Ink4a or C/EBPδ expression occurs stochastically in a subpopulation of cells resulting in their senescence. In this model, the capacity for self-renewal of the noncommitted cells is not affected. (b) Threshold model: expression of p16Ink4a or C/EBPδ increases or decreases, respectively, uniformly in the tissue specific. Self-renewing cells compartment is compromised over time. Self-renewal is indicated by curved arrows; committed cells are colored blue.
Addressing this question has important implications for future “antiaging” therapies or “antidifferentiation” therapies. The model in which cell-cycle entry is decreased (threshold model) suggests that the age-induced defects in proliferation or the hormone-induced defects in proliferation could be improved merely by reducing p16Ink4a levels or C/EBPδ levels or otherwise increasing cdk4/6 activity in these cells. The commitment model, however, suggests that the defects in self-renewal could only be remedied through more drastic self-renewing cells from an exogenous source. The understanding of the in vivo regulation of the Ink4a/Arf/Ink4b locus with aging (oxidative stress) or C/EBPδ locus with differentiation is needed further.
Acknowledgments
The authors thank Drs. K. Itakura, G. Gachelin, R. Chiu, R. Eckner, P.-T. Yao, T. Kouzarides, C. D. Allis, D. M. Livingston, and M. Horikoshi for their helpful discussions and reagents. This work was supported by Grants from KMU COE (no. KMU-EM-99-3 to E-M. Tsai and K. K. Yokoyama), the Department of health, Executive Yuan, Taiwan (no. DOH99-TD-C-111-002 to M-F. Hou), and the Ministry of Education, Culture, Sports and Technology of Japan (to N. Yamaguchi and K. K. Yokoyama).
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annual Review of Biochemistry. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 3.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Ekwall K. Genome-wide analysis of HDAC function. Trends in Genetics. 2005;21(11):608–615. doi: 10.1016/j.tig.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 6.Andrews AJ, Chen X, Zevin A, et al. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnuclesomal histone DNA interactions. Molecular Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmad K, Henikoff S. Epigenetic consequences of nucleosome dynamics. Cell. 2002;111(3):281–284. doi: 10.1016/s0092-8674(02)01081-4. [DOI] [PubMed] [Google Scholar]
- 8.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140(2):183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Current Opinion in Genetics and Development. 2005;15(2):163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Svejstrup JQ. Transcription, histones face the FACT. Science. 2003;301(5636):1053–1055. doi: 10.1126/science.1088901. [DOI] [PubMed] [Google Scholar]
- 11.Workman JL. Nucleosome displacement in transcription. Genes and Development. 2006;20(15):2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 12.Akey CW, Luger K. Histone chaperones and nucleosome assembly. Current Opinion in Structural Biology. 2003;13(1):6–14. doi: 10.1016/s0959-440x(03)00002-2. [DOI] [PubMed] [Google Scholar]
- 13.Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochimica et Biophysica Acta. 2004;1677(1–3):3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Ray-Gallet D, Quivy J-P, Scamps C, Martini EM-D, Lipinski M, Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Molecular Cell. 2002;9(5):1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- 15.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116(1):51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 16.Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein- protein interactions. Molecular and Cellular Biology. 1997;17(6):3094–3102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin C, Ugai H, Song J, et al. Identification of mouse Jun dimerization protein 2 as a novel repressor of ATF-2. FEBS Letters. 2001;489(1):34–41. doi: 10.1016/s0014-5793(00)02387-5. [DOI] [PubMed] [Google Scholar]
- 18.Broder YC, Katz S, Aronheim A. The Ras recruitment system, a novel approach to the study of protein-protein interactions. Current Biology. 1998;8(20):1121–1124. doi: 10.1016/s0960-9822(98)70467-1. [DOI] [PubMed] [Google Scholar]
- 19.Wardell SE, Boonyaratanakornkit V, Adelman JS, Aronheim A, Edwards DP. Jun dimerization protein 2 functions as a progesterone receptor N-terminal domain coactivator. Molecular and Cellular Biology. 2002;22(15):5451–5466. doi: 10.1128/MCB.22.15.5451-5466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan J, Jin C, Murata T, Yokoyama KK. Sequence specific transcription factor, JDP2 interacts with histone and inhibits p300-mediated histone acetylation. Nucleic Acids Research. Supplement. 2003;(3):305–306. doi: 10.1093/nass/3.1.305. [DOI] [PubMed] [Google Scholar]
- 21.Katz S, Aronheim A. Differential targeting of the stress mitogen-activated protein kinases to the c-Jun dimerization protein 2. Biochemical Journal. 2002;368(3):939–945. doi: 10.1042/BJ20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz S, Heinrich R, Aronheim A. The AP-1 repressor, JDP2, is a bona fide substrate for the c-Jun N-terminal kinase. FEBS Letters. 2001;506(3):196–200. doi: 10.1016/s0014-5793(01)02907-6. [DOI] [PubMed] [Google Scholar]
- 23.Kimura M. IRF2-binding protein-1 is a JDP2 ubiquitin ligase and an inhibitor of ATF2-dependent transcription. FEBS Letters. 2008;582(19):2833–2837. doi: 10.1016/j.febslet.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 24.Piu F, Aronheim A, Katz S, Karin M. AP-1 repressor protein JDP-2: inhibition of UV-mediated apoptosis through p53 down-regulation. Molecular and Cellular Biology. 2001;21(9):3012–3024. doi: 10.1128/MCB.21.9.3012-3024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blazek E, Wasmer S, Kruse U, Aronheim A, Aoki M, Vogt PK. Partial oncogenic transformation of chicken embryo fibroblasts by Jun dimerization protein 2, a negative regulator of TRE- and CRE-dependent transcription. Oncogene. 2003;22(14):2151–2159. doi: 10.1038/sj.onc.1206312. [DOI] [PubMed] [Google Scholar]
- 26.Holding C. JDP2—a cell-cycle master switch? Trends in Biochemical Sciences. 2002;27(12):p. 603. [Google Scholar]
- 27.Ostrovsky O, Bengal E, Aronheim A. Induction of terminal differentiation by the c-Jun dimerization protein JDP2 in C2 myoblasts and rhabdomyosarcoma cells. Journal of Biological Chemistry. 2002;277(42):40043–40054. doi: 10.1074/jbc.M205494200. [DOI] [PubMed] [Google Scholar]
- 28.Kawaida R, Ohtsuka T, Okutsu J, et al. Jun dimerization protein 2 (JDP2), a member of the AP-1 family of transcription factor, mediates osteoclast differentiation induced by RANKL. Journal of Experimental Medicine. 2003;197(8):1029–1035. doi: 10.1084/jem.20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang HC, Martins CP, Bronkhorst Y, et al. Identification of oncogenes collaborating with p27Kip1 loss by insertional mutagenesis and high-throughput insertion site analysis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(17):11293–11298. doi: 10.1073/pnas.162356099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinrich R, Livne E, Ben-Izhak O, Aronheim A. The c-Jun dimerization protein 2 inhibits cell transformation and acts as a tumor suppressor gene. Journal of Biological Chemistry. 2004;279(7):5708–5715. doi: 10.1074/jbc.M307608200. [DOI] [PubMed] [Google Scholar]
- 31.Jin C, Li H, Murata T, et al. JDP2, a repressor of AP-1, recruits a histone deacetylase 3 complex to inhibit the retinoic acid-induced differentiation of F9 cells. Molecular and Cellular Biology. 2002;22(13):4815–4826. doi: 10.1128/MCB.22.13.4815-4826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin C, Kato K, Chimura T, et al. Regulation of histone acetylation and nucleosome assembly by transcription factor JDP2. Nature Structural and Molecular Biology. 2006;13(4):331–338. doi: 10.1038/nsmb1063. [DOI] [PubMed] [Google Scholar]
- 33.Nakade K, Pan J, Yoshiki A, et al. JDP2 suppresses adipocyte differentiation by regulating histone acetylation. Cell Death and Differentiation. 2007;14(8):1398–1405. doi: 10.1038/sj.cdd.4402129. [DOI] [PubMed] [Google Scholar]
- 34.Yang X-J, Ogryzko VV, Nishikawa J-I, Howard BH, Nakatani Y. A p300/CPB-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382(6589):319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 35.Kawasaki H, Schiltz L, Chiu R, et al. ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature. 2000;405(6783):195–200. doi: 10.1038/35012097. [DOI] [PubMed] [Google Scholar]
- 36.Karanam B, Wang L, Wang D, et al. Multiple roles for acetylation in the interaction of p300 HAT with ATF-2. Biochemistry. 2007;46(28):8207–8216. doi: 10.1021/bi7000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruhat A, Chrérasse Y, Maurin A-C, et al. ATF2 is required for amino acid-regulated transcription by orchestrating specific histone acetylation. Nucleic Acids Research. 2007;35(4):1312–1321. doi: 10.1093/nar/gkm038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo S-B, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104(1):119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 39.Seo S-B, Macfarlan T, McNamara P, et al. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. Journal of Biological Chemistry. 2002;277(16):14005–14010. doi: 10.1074/jbc.M112455200. [DOI] [PubMed] [Google Scholar]
- 40.Nakade K, Pan J, Yamasaki T, Murata T, Wasylyk B, Yokoyama KK. JDP2 (Jun dimerization protein 2)-deficient mouse embryonic fibroblasts are resistant to replicative senescene. Journal of Biological Chemistry. 2009;284(16):10808–10817. doi: 10.1074/jbc.M808333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kehat I, Hasin T, Aronheim A. The role of basic leucine zipper protein-mediated transcription in physiological and pathological myocardial hypertrophy. Annals of the New York Academy of Sciences. 2006;1080:97–109. doi: 10.1196/annals.1380.009. [DOI] [PubMed] [Google Scholar]
- 42.Rajala MW, Scherer PE. Minireview: the adipocyte—at the crossroads of energy homeostasis, inflammation, and atherosclerosis. Endocrinology. 2003;144(9):3765–3773. doi: 10.1210/en.2003-0580. [DOI] [PubMed] [Google Scholar]
- 43.Kahn BB, Flier JS. Obesity and insulin resistance. Journal of Clinical Investigation. 2000;106(4):473–481. doi: 10.1172/JCI10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rangwala SM, Lazar MA. Transcriptional control of adipogenesis. Annual Review of Nutrition. 2000;20:535–559. doi: 10.1146/annurev.nutr.20.1.535. [DOI] [PubMed] [Google Scholar]
- 45.Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annual Review of Cell and Developmental Biology. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO Journal. 1997;16(24):7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans RM, Barish GD, Wang Y-X. PPARs and the complex journey to obesity. Nature Medicine. 2004;10(4):355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 48.Morrison RF, Farmer SR. Hormonal signaling and transcriptional control of adipocyte differentiation. Journal of Nutrition. 2000;130(12):3116S–3121S. doi: 10.1093/jn/130.12.3116S. [DOI] [PubMed] [Google Scholar]
- 49.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes and Development. 2000;14(11):1293–1307. [PubMed] [Google Scholar]
- 50.Lin F-T, MacDougald OA, Diehl AM, Lane MD. A 30-kDa alternative translation product of the CCAAT/enhancer binding protein α message: transcriptional activator lacking antimitotic activity. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(20):9606–9610. doi: 10.1073/pnas.90.20.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christy RJ, Kaestner KH, Geiman DE, Lane MD. CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(6):2593–2597. doi: 10.1073/pnas.88.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Z, Xie Y, Bucher NLR, Farmer SR. Conditional ectopic expression of C/EBPβ in NIH-3T3 cells induces PPARγ and stimulates adipogenesis. Genes and Development. 1995;9(19):2350–2363. doi: 10.1101/gad.9.19.2350. [DOI] [PubMed] [Google Scholar]
- 53.Fajas L, Schoonjans K, Gelman L, et al. Regulation of peroxisome proliferator-activated receptor γ expression by adipocyte differentiation and determination factor 1/sterol regulatory element binding protein 1: implications for adipocyte differentiation and metabolism. Molecular and Cellular Biology. 1999;19(8):5495–5503. doi: 10.1128/mcb.19.8.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shao D, Lazar MA. Peroxisome proliferator activated receptor γ, CCAAT/enhancer-binding protein α, and cell cycle status regulate the commitment to adipocyte differentiation. Journal of Biological Chemistry. 1997;272(34):21473–21478. doi: 10.1074/jbc.272.34.21473. [DOI] [PubMed] [Google Scholar]
- 55.Schwarz EJ, Reginato MJ, Shao D, Krakow SL, Lazar MA. Retinoic acid blocks adipogenesis by inhibiting C/EBPβ-mediated transcription. Molecular and Cellular Biology. 1997;17(3):1552–1561. doi: 10.1128/mcb.17.3.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Z, Rosen ED, Brun R, et al. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Molecular Cell. 1999;3(2):151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- 57.Kang S, Bajnok L, Longo KA, et al. Effects of Wnt signaling on brown adipocyte differentiation and metabolism mediated by PGC-1α. Molecular and Cellular Biology. 2005;25(4):1272–1282. doi: 10.1128/MCB.25.4.1272-1282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hansen JB, Jørgensen C, Petersen RK, et al. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(12):4112–4117. doi: 10.1073/pnas.0301964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guan H-P, Ishizuka T, Chui PC, Lehrke M, Lazar MA. Corepressors selectively control the transcriptional activity of PPARγ in adipocytes. Genes and Development. 2005;19(4):453–461. doi: 10.1101/gad.1263305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sherr CJ. Cancer cell cycles. Science. 1996;274(5293):1672–1674. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 61.Weinberg RA. E2F and cell proliferation: a world turned upside down. Cell. 1996;85(4):457–459. doi: 10.1016/s0092-8674(00)81244-1. [DOI] [PubMed] [Google Scholar]
- 62.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes and Development. 2000;14(19):2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 63.Harbour JW, Dean DC. Rb function in cell-cycle regulation and apoptosis. Nature Cell Biology. 2000;2(4):E65–E67. doi: 10.1038/35008695. [DOI] [PubMed] [Google Scholar]
- 64.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes and Development. 1999;13(12):1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 65.Pavletich NP. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. Journal of Molecular Biology. 1999;287(5):821–828. doi: 10.1006/jmbi.1999.2640. [DOI] [PubMed] [Google Scholar]
- 66.Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutation Research. 2005;576(1-2):22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 67.El-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 68.Hermeking H, Lengauer C, Polyak K, et al. 14-3-3σ is a p53-regulated inhibitor of G2/M progression. Molecular Cell. 1997;1(1):3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 69.Zhan Q, Chen I-T, Antinore MJ, Fornace AJ., Jr. Tumor suppressor p53 can participate in transcriptional induction of the GADD45 promoter in the absence of direct DNA binding. Molecular and Cellular Biology. 1998;18(5):2768–2778. doi: 10.1128/mcb.18.5.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80(2):293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 71.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Molecular Cell. 2001;7(3):683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 72.Nagata S. Apoptosis by death factor. Cell. 1997;88(3):355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 73.Wu GS, Burns TF, McDonald ER, III, et al. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nature Genetics. 1997;17(2):141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 74.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Molecular Cell. 2001;7(3):673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 75.Reisman D, Loging WT. Transcriptional regulation of the p53 tumor suppressor gene. Seminars in Cancer Biology. 1998;8(5):317–324. doi: 10.1006/scbi.1998.0094. [DOI] [PubMed] [Google Scholar]
- 76.Fu L, Benchimol S. Participation of the human p53 3′UTR in translational repression and activation following γ-irradiation. EMBO Journal. 1997;16(13):4117–4125. doi: 10.1093/emboj/16.13.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Letters. 1997;420(1):25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 78.Perry ME, Piette J, Zawadzki JA, Harvey D, Levine AJ. The mdm-2 gene is induced in response to UV light in a p53-dependent manner. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(24):11623–11627. doi: 10.1073/pnas.90.24.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO Journal. 1999;18(1):22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91(3):325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 81.Mayo LD, Turchi JJ, Berberich SJ. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Research. 1997;57(22):5013–5016. [PubMed] [Google Scholar]
- 82.Khanna KK, Keating KE, Kozlov S, et al. ATM associates with and phosphorylates p53: mapping the region of interaction. Nature Genetics. 1998;20(4):398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 83.Banin S, Moyal L, Shieh S-Y, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281(5383):1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 84.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nature Reviews Cancer. 2006;6(9):663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 85.Van Maerken T, Vandesompele J, Rihani A, De Paepe A, Speleman F. Escape from p53-mediated tumor surveillance in neuroblastoma: switching off the p14ARF-MDM2-p53 axis. Cell Death and Differentiation. 2009;16(12):1563–1572. doi: 10.1038/cdd.2009.138. [DOI] [PubMed] [Google Scholar]
- 86.Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85(1):27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 87.Bodnar AG, Ouellette M, Frolkis M, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279(5349):349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 88.Lee H-W, Blasco MA, Gottlieb GJ, Horner JW, II, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392(6676):569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 89.Blasco MA, Lee H-W, Hande MP, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91(1):25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 90.García-Cao I, García-Cao M, Tomás-Loba A, et al. Increased p53 activity does not accelerate telomere-driven ageing. EMBO Reports. 2006;7(5):546–552. doi: 10.1038/sj.embor.7400667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ressler S, Bartkova J, Niederegger H, et al. p16INK4A is a robust in vivo biomarker of cellular aging in human skin. Aging Cell. 2006;5(5):379–389. doi: 10.1111/j.1474-9726.2006.00231.x. [DOI] [PubMed] [Google Scholar]
- 92.Efeyan A, Garcia-Cao I, Herranz D, Velasco-Miguel S, Serrano M. Tumour biology: policing of oncogene activity by p53. Nature. 2006;443(7108):p. 159. doi: 10.1038/443159a. [DOI] [PubMed] [Google Scholar]
- 93.Mallette FA, Gaumont-Leclerc M-F, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes and Development. 2007;21(1):43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 95.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 98.Melzer D, Frayling TM, Murray A, et al. A common variant of the p16INK4a genetic region is associated with physical function in older people. Mechanisms of Ageing and Development. 2007;128(5-6):370–377. doi: 10.1016/j.mad.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mason PJ, Bessler M. Heterozygous telomerase deficiency in mouse and man: when less is definitely not more. Cell Cycle. 2004;3(9):1127–1129. [PubMed] [Google Scholar]
- 101.Vitolo JM, Thiriet C, Hayes JJ. The H3-H4 N-terminal tail domains are the primary mediators of transcription factor IIIA access to 5S DNA within a nucleosome. Molecular and Cellular Biology. 2000;20(6):2167–2175. doi: 10.1128/mcb.20.6.2167-2175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang Z, Zheng C, Thiriet C, Hayes JJ. The core histone N-terminal tail domains negatively regulate binding of transcription factor IIIA to a nucleosome containing a 5S RNA gene via a novel mechanism. Molecular and Cellular Biology. 2005;25(1):241–249. doi: 10.1128/MCB.25.1.241-249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li F, Macfarlan T, Pittman RN, Chakravarti D. Ataxin-3 is a histone-binding protein with two independent transcriptional corepressor activities. Journal of Biological Chemistry. 2002;277(47):45004–45012. doi: 10.1074/jbc.M205259200. [DOI] [PubMed] [Google Scholar]
- 104.Yu J, Li Y, Ishizuka T, et al. A SANT motif in the SMRT corepressor in triplets the histone code and promotes histone acetylations. The EMBO Journal. 2003;22:3403–3410. doi: 10.1093/emboj/cdg326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choi B, Ko JK, Shin J. The transcriptional corepressor, PELP1, recruits HDAC2 and masks histones using two separate domains. Journal of Biological Chemistry. 2004;279(49):50930–50941. doi: 10.1074/jbc.M406831200. [DOI] [PubMed] [Google Scholar]
- 106.Loven MA, Muster N, Yates JR, et al. A novel estrogen receptor alpha-associated pasortien, template-activation factor I beta, inhibits acetylation and transactivation. Molecular Endocrinology. 2003;17:67–78. doi: 10.1210/me.2002-0280. [DOI] [PubMed] [Google Scholar]
- 107.Loven MA, Davis RE, Curtis CD, Muster N, Yates JR, Nardulli AM. A novel estrogen receptor α-associated protein alters receptor-deoxyribonucleic acid interactions and represses receptor-mediated transcription. Molecular Endocrinology. 2004;18(11):2649–2659. doi: 10.1210/me.2003-0195. [DOI] [PubMed] [Google Scholar]
- 108.Macfarlan T, Kutney S, Altman B, Montross R, Yu J, Chakravarti D. Human THAP7 is a chromatin-associated, histone tail-binding protein that represses transcription via recruitment of HDAC3 and nuclear hormone receptor corepressor. Journal of Biological Chemistry. 2005;280(8):7346–7358. doi: 10.1074/jbc.M411675200. [DOI] [PubMed] [Google Scholar]
- 109.Macfarlan T, Parker JB, Nagata K, Chakravarti D. Thanatos-associated protein 7 associates with template activating factor-Iβ and inhibits histone acetylation to repress transcription. Molecular Endocrinology. 2006;20(2):335–347. doi: 10.1210/me.2005-0248. [DOI] [PubMed] [Google Scholar]
- 110.Hublitz P, Kunowska N, Mayer UP, et al. NIR is a novel INHAT repressor that modulates the transcriptional activity of p53. Genes and Development. 2005;19(23):2912–2924. doi: 10.1101/gad.351205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hong W, Kim AY, Ky S, et al. Inhibition of CBP-mediated protein acetylation by the Ets family oncoprotein PU.1. Molecular and Cellular Biology. 2002;22(11):3729–3743. doi: 10.1128/MCB.22.11.3729-3743.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO Journal. 2005;24(21):3712–3723. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hong R, Chakravarti D. The human proliferating cell nuclear antigen regulates transcriptional coactivator p300 activity and promotes transcriptional repression. Journal of Biological Chemistry. 2003;278(45):44505–44513. doi: 10.1074/jbc.M303138200. [DOI] [PubMed] [Google Scholar]
- 114.Suzuki T, Muto S, Miyamoto S, Aizawa K, Horikoshi M, Nagai R. Functional interaction of the DNA-binding transcription factor Sp1 through its DNA-binding domain with the histone chaperone TAF-I. Journal of Biological Chemistry. 2003;278(31):28758–28764. doi: 10.1074/jbc.M302228200. [DOI] [PubMed] [Google Scholar]
- 115.Miyamoto S, Suzuki T, Muto S, et al. Positive and negative regulation of the cardiovascular transcription actor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Molecular and Cellular Biology. 2003;23:8528–8541. doi: 10.1128/MCB.23.23.8528-8541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Angelov D, Lenouvel F, Hans F, et al. The histone octamer is invisible when NF-κB binds to the nucleosome. Journal of Biological Chemistry. 2004;279(41):42374–42382. doi: 10.1074/jbc.M407235200. [DOI] [PubMed] [Google Scholar]
- 117.Mo X, Kowenz-Leutz E, Laumonnier Y, Xu H, Leutz A. Histone H3 tail positioning and acetylation by the c-Myb but not the v-Myb DNA-binding SANT domain. Genes and Development. 2005;19(20):2447–2457. doi: 10.1101/gad.355405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. Journal of Cell Biology. 2007;176(6):757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Agelopoulos M, Thanos D. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO Journal. 2006;25(20):4843–4853. doi: 10.1038/sj.emboj.7601364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pan J, Nakade K, Huang Y-C, et al. Suppression of cell cycle progression by Jun dimerization protein (JDP2) involves down-regulation of cyclin A2. Oncogene. 2010 doi: 10.1038/onc.2010.355. [DOI] [PMC free article] [PubMed] [Google Scholar]