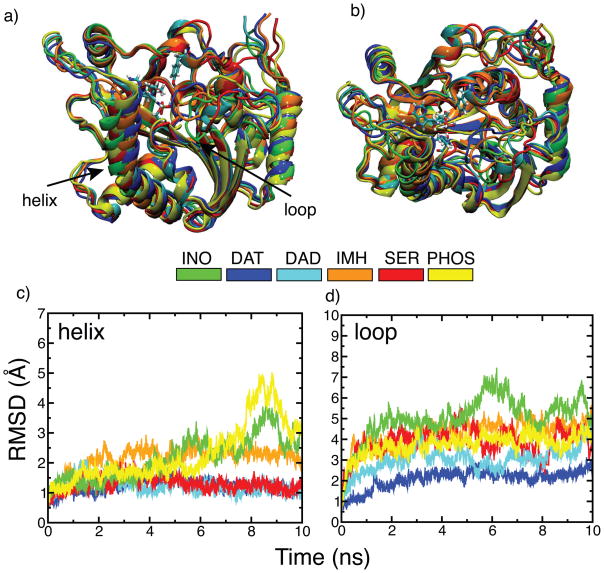
Figure 5.
Dynamical changes observed in the helix and loop residues of hPNP homotrimer during 10 ns simulations in the presence of different ligands. a) Side-view of one of the chains for each inhibitor bound structure superimposed with respect to the INO simulated structure (green). Backbone atoms of the average simulated structures obtained from the last 500 ps of 10 ns dynamics runs were used for superimposition. b) Top-view of the same superimposed structures. c) Backbone RMSD of only helix residues (256–268) of all three chains of the hPNP homotrimer bound to different ligands after aligning strutures on all backbone atoms of the hPNP. d) Backbone RMSD of only loop residues (59–68) for all three chains of the hPNP homotrimer after aligning structures on all backbone atoms of the hPNP. RMSD of the helix and loop segments for the individual chains of all complexes is available in the supporting information Fig. S1.