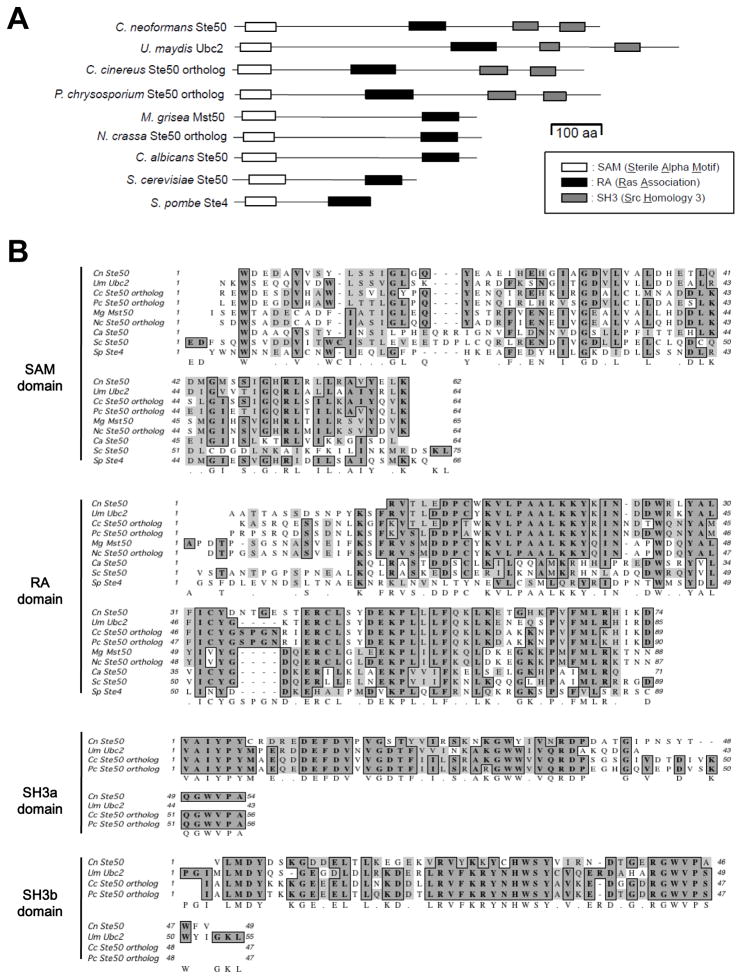
Fig. 1.
Comparison of Ste50 orthologs between C. neoformans and other fungi. (A) Each Ste50 ortholog diagram shows functional protein domains, which were identified by the Pfam 24.0 database (http://pfam.sanger.ac.uk/). Each domain indicates the following: SAM, sterile alpha motif; RA, Ras- associated; SH3, Src homology 3. (B) Multiple sequence alignment of Ste50 orthologs is depicted by Clustal W alignment from MacVector software (versions 7.2.3, Accelrys). Protein sequences of Ste50 orthologs were retrieved from the following database: C. neoformans Ste50 - CNAG_07507 from the C. neoformans var. grubii H99 database of the Broad Institute (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html); Ustilago maydis Ubc2 - GenBank accession number AAK4932; Coprinus cinereus - locus CC1G_00975.3 from the C. cinereus Sequencing Project Database of the Broad Institute. (http://www.broadinstitute.org/annotation/genome/coprinus_cinereus/MultiHome.html); Phanerochaete chrysosporium Ste50 ortholog - protein ID 2321 at locus Phchr1/scaffold_3:1012644-101605 from the sequencing data produced by the United States Department of Energy, Joint Genome Institute (http://genome.jgi-psf.org/Phchr1/Phchr1.home.html); Neurospora crassa Ste50 ortholog - GenBank accession number XP_956774; Magnaporthe grisae Mst50 - GenBank accession number XP_359578; Candida albicans Ste50 - GenBank accession number XP_721713; Saccharomyces cerevisiae Ste50 - GenBank accession number NP_009898; Schizosaccharomyces pombe Ste4; GenBank accession number CAB38684.