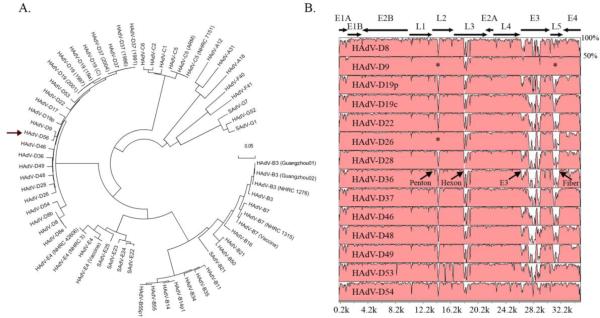
Fig. 1.
Whole genome analysis of HAdV-D56. (A) Bootstrap- confirmed neighbor-joining tree designed from MEGA 4.0.2 demonstrates relationships between HAdV-D56 and all other completely sequenced adenovirus genomes. (arrow: HAdV-D56). Bootstrap values for the species nodes were 100. The bootstrap value for the HAdV-D56 and HAdV-D9 node was also 100. (B) Global pairwise sequence comparison of HAdV-D56 with fourteen other completely sequenced HAdV-D types. Percent sequence conservation is reflected in the height of each data point along the y-axis. The penton base, hexon, E3, and fiber protein coding regions are divergent within species D, except for the penton base and fiber regions of HAdV-D9 and HAdV-D26 (*).