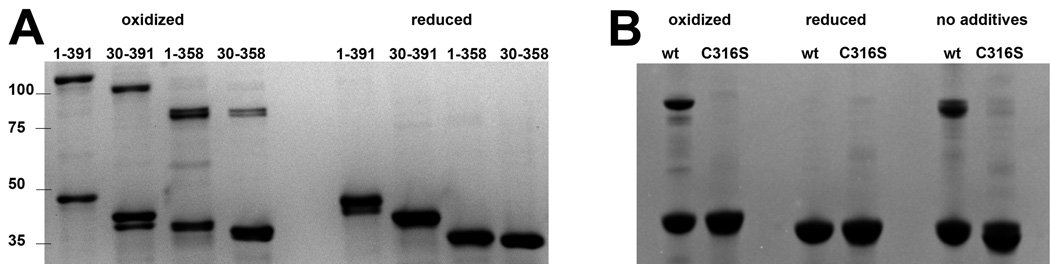
Fig. 3.
SDS PAGE analysis of oxidized and reduced UvsX. (A) Wild type full length and truncated UvsX proteins show partial dimerization when oxidized and a single band when reduced. The N-terminal deletion mutant UvsX30–391 shows degradation when oxidized, but the reduced sample runs as a single species. The C-terminal deletion mutant UvsX1–358 and the core domain UvsX30–358 migrate similarly on the gel. The band positions of protein standards are marked with their molecular weights (kDa) on the left. Note that the dimeric fractions of both samples which lack the disordered acidic C-terminal region run at a molecular weight more consistent with the calculated molecular weight. The calculated molecular weight values for each dimer are: 93 kDa for UvsX1–391, 86 kDa for UvsX30–391, 84 kDa for UvsX1–358 and 80 kDa for UvsX30–358. (B) The C316S point mutant of full-length UvsX fails to dimerize in oxidizing conditions. Wild type (wt) UvsX exhibits dimerization in the loading buffer without addition of oxidizing agents (right hand side). Reduced UvsX protein was supplemented with 5 mM TCEP, and oxidized protein with 1% H2O2.